Housekeeping genes essential for pantothenate biosynthesis are plasmid-encoded in Rhizobium etli and Rhizobium leguminosarum
- PMID: 21463532
- PMCID: PMC3082293
- DOI: 10.1186/1471-2180-11-66
Housekeeping genes essential for pantothenate biosynthesis are plasmid-encoded in Rhizobium etli and Rhizobium leguminosarum
Abstract
Background: A traditional concept in bacterial genetics states that housekeeping genes, those involved in basic metabolic functions needed for maintenance of the cell, are encoded in the chromosome, whereas genes required for dealing with challenging environmental conditions are located in plasmids. Exceptions to this rule have emerged from genomic sequence data of bacteria with multipartite genomes. The genome sequence of R. etli CFN42 predicts the presence of panC and panB genes clustered together on the 642 kb plasmid p42f and a second copy of panB on plasmid p42e. They encode putative pantothenate biosynthesis enzymes (pantoate-β-alanine ligase and 3-methyl-2-oxobutanoate hydroxymethyltransferase, respectively). Due to their ubiquitous distribution and relevance in the central metabolism of the cell, these genes are considered part of the core genome; thus, their occurrence in a plasmid is noteworthy. In this study we investigate the contribution of these genes to pantothenate biosynthesis, examine whether their presence in plasmids is a prevalent characteristic of the Rhizobiales with multipartite genomes, and assess the possibility that the panCB genes may have reached plasmids by horizontal gene transfer.
Results: Analysis of mutants confirmed that the panC and panB genes located on plasmid p42f are indispensable for the synthesis of pantothenate. A screening of the location of panCB genes among members of the Rhizobiales showed that only R. etli and R. leguminosarum strains carry panCB genes in plasmids. The panCB phylogeny attested a common origin for chromosomal and plasmid-borne panCB sequences, suggesting that the R. etli and R. leguminosarum panCB genes are orthologs rather than xenologs. The panCB genes could not totally restore the ability of a strain cured of plasmid p42f to grow in minimal medium.
Conclusions: This study shows experimental evidence that core panCB genes located in plasmids of R. etli and R. leguminosarum are indispensable for the synthesis of pantothenate. The unusual presence of panCB genes in plasmids of Rhizobiales may be due to an intragenomic transfer from chromosome to plasmid. Plasmid p42f encodes other functions required for growth in minimal medium. Our results support the hypothesis of cooperation among different replicons for basic cellular functions in multipartite rhizobia genomes.
Figures
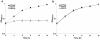
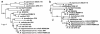

References
-
- Romero D, Brom S. In: Plasmid biology. Phillips G, Funnell BE, editor. Washington, D.C: American Society for Microbiology; 2004. The symbiotic plasmids of the Rhizobiaceae; pp. 271–290.
-
- Young JP, Crossman LC, Johnston AW, Thomson NR, Ghazoui ZF, Hull KH, Wexler M, Curson AR, Todd JD, Poole PS, Mauchline TH, East AK, Quail MA, Churcher C, Arrowsmith C, Cherevach I, Chillingworth T, Clarke K, Cronin A, Davis P, Fraser A, Hance Z, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, Whitehead S, Parkhill J. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006;7:R34. doi: 10.1186/gb-2006-7-4-r34. - DOI - PMC - PubMed
-
- Crossman LC, Castillo-Ramírez S, McAnnula C, Lozano L, Vernikos GS, Acosta JL, Ghazoui ZF, Hernández-González I, Meakin G, Walker AW, Hynes MF, Young JPW, Downie JA, Romero D, Johnston AWB, Dávila G, Parkhill J, González V. A common genomic framework for a diverse assembly of plasmids in the symbiotic nitrogen fixing bacteria. PLoS ONE. 2007;3:e2567. doi: 10.1371/journal.pone.0002567. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

