Super-resolution imaging of the nucleoid-associated protein HU in Caulobacter crescentus
- PMID: 21463569
- PMCID: PMC3072666
- DOI: 10.1016/j.bpj.2011.02.022
Super-resolution imaging of the nucleoid-associated protein HU in Caulobacter crescentus
Abstract
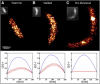
Little is known about the structure and function of most nucleoid-associated proteins (NAPs) in bacteria. One reason for this is that the distribution and structure of the proteins is obfuscated by the diffraction limit in standard wide-field and confocal fluorescence imaging. In particular, the distribution of HU, which is the most abundant NAP, has received little attention. In this study, we investigate the distribution of HU in Caulobacter crescentus using a combination of super-resolution fluorescence imaging and spatial point statistics. By simply increasing the laser power, single molecules of the fluorescent protein fusion HU2-eYFP can be made to blink on and off to achieve super-resolution imaging with a single excitation source. Through quantification by Ripley's K-test and comparison with Monte Carlo simulations, we find the protein is slightly clustered within a mostly uniform distribution throughout the swarmer and stalked stages of the cell cycle but more highly clustered in predivisional cells. The methods presented in this letter should be of broad applicability in the future study of prokaryotic NAPs.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

