Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa
- PMID: 21463573
- PMCID: PMC3072661
- DOI: 10.1016/j.bpj.2011.02.020
Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa
Abstract
Bacterial biofilms are structured multicellular communities that are responsible for a broad range of infections. Knowing how free-swimming bacteria adapt their motility mechanisms near a surface is crucial for understanding the transition from the planktonic to the biofilm phenotype. By translating microscopy movies into searchable databases of bacterial behavior and developing image-based search engines, we were able to identify fundamental appendage-specific mechanisms for the surface motility of Pseudomonas aeruginosa. Type IV pili mediate two surface motility mechanisms: horizontally oriented crawling, by which the bacterium moves lengthwise with high directional persistence, and vertically oriented walking, by which the bacterium moves with low directional persistence and high instantaneous velocity, allowing it to rapidly explore microenvironments. The flagellum mediates two additional motility mechanisms: near-surface swimming and surface-anchored spinning, which often precedes detachment from a surface. Flagella and pili interact cooperatively in a launch sequence whereby bacteria change orientation from horizontal to vertical and then detach. Vertical orientation facilitates detachment from surfaces and thereby influences biofilm morphology.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


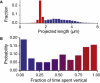


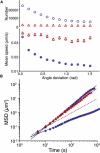
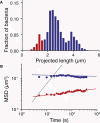
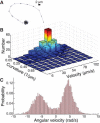

References
-
- Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. - PubMed
-
- Anderson G.G., O'Toole G.A. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 2008;322:85–105. - PubMed
-
- Hall-Stoodley L., Stoodley P. Evolving concepts in biofilm infections. Cell. Microbiol. 2009;11:1034–1043. - PubMed
-
- Nassif X., Marceau M., Taha M.K. Type-4 pili and meningococcal adhesiveness. Gene. 1997;192:149–153. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

