EOBII controls flower opening by functioning as a general transcriptomic switch
- PMID: 21464473
- PMCID: PMC3177291
- DOI: 10.1104/pp.111.176248
EOBII controls flower opening by functioning as a general transcriptomic switch
Abstract
R2R3-MYB transcription factors (TFs) are involved in diverse aspects of plant biology. Recently an R2R3-MYB was identified in Petunia x hybrida line P720 to have a role in the transcriptional regulation of floral volatile production. We propose a more foundational role for the R2R3-MYB TF EMISSION OF BENZENOIDS II (EOBII). The homolog of EOBII was isolated and characterized from P. x hybrida 'Mitchell Diploid' (MD) and Nicotiana attenuata. For both MD and N. attenuata, EOBII transcript accumulates to high levels in floral tissue with maximum accumulation at flower opening. When EOBII transcript levels are severely reduced using a stable RNAi (ir) approach in MD and N. attenuata, ir-EOBII flowers fail to enter anthesis and prematurely senesce. Transcript accumulation analysis demonstrated core phenylpropanoid pathway transcripts and cell wall modifier transcript levels are altered in ir-EOBII flowers. These flowers can be partially complemented by feeding with a sucrose, t-cinnamic acid, and gibberellic acid solution; presumably restoring cellular aspects sufficient for flower opening. Additionally, if ethylene sensitivity is blocked in either MD or N. attenuata, ir-EOBII flowers enter anthesis. These experiments demonstrate one R2R3-MYB TF can control a highly dynamic process fundamental to sexual reproduction in angiosperms: the opening of flowers.
Figures

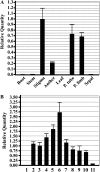







References
-
- Ashe HL, Briscoe J. (2006) The interpretation of morphogen gradients. Development 133: 385–394 - PubMed
-
- Ben-Nissan G, Weiss D. (1996) The petunia homologue of tomato gast1: transcript accumulation coincides with gibberellin-induced corolla cell elongation. Plant Mol Biol 32: 1067–1074 - PubMed
-
- Coen ES, Meyerowitz EM. (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

