Molecular recognition of H3/H4 histone tails by the tudor domains of JMJD2A: a comparative molecular dynamics simulations study
- PMID: 21464980
- PMCID: PMC3064570
- DOI: 10.1371/journal.pone.0014765
Molecular recognition of H3/H4 histone tails by the tudor domains of JMJD2A: a comparative molecular dynamics simulations study
Abstract
Background: Histone demethylase, JMJD2A, specifically recognizes and binds to methylated lysine residues at histone H3 and H4 tails (especially trimethylated H3K4 (H3K4me3), trimethylated H3K9 (H3K9me3) and di,trimethylated H4K20 (H4K20me2, H4K20me3)) via its tandem tudor domains. Crystal structures of JMJD2A-tudor binding to H3K4me3 and H4K20me3 peptides are available whereas the others are not. Complete picture of the recognition of the four histone peptides by the tandem tudor domains yet remains to be clarified.
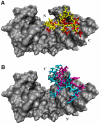
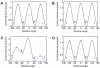
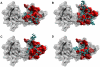
Methodology/principal findings: We report a detailed molecular dynamics simulation and binding energy analysis of the recognition of JMJD2A-tudor with four different histone tails. 25 ns fully unrestrained molecular dynamics simulations are carried out for each of the bound and free structures. We investigate the important hydrogen bonds and electrostatic interactions between the tudor domains and the peptide molecules and identify the critical residues that stabilize the complexes. Our binding free energy calculations show that H4K20me2 and H3K9me3 peptides have the highest and lowest affinity to JMJD2A-tudor, respectively. We also show that H4K20me2 peptide adopts the same binding mode with H4K20me3 peptide, and H3K9me3 peptide adopts the same binding mode with H3K4me3 peptide. Decomposition of the enthalpic and the entropic contributions to the binding free energies indicate that the recognition of the histone peptides is mainly driven by favourable van der Waals interactions. Residue decomposition of the binding free energies with backbone and side chain contributions as well as their energetic constituents identify the hotspots in the binding interface of the structures.
Conclusion: Energetic investigations of the four complexes suggest that many of the residues involved in the interactions are common. However, we found two receptor residues that were related to selective binding of the H3 and H4 ligands. Modifications or mutations on one of these residues can selectively alter the recognition of the H3 tails or the H4 tails.
Conflict of interest statement
Figures




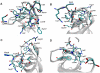
Similar articles
-
Engineering human JMJD2A tudor domains for an improved understanding of histone peptide recognition.Proteins. 2023 Jan;91(1):32-46. doi: 10.1002/prot.26408. Epub 2022 Aug 16. Proteins. 2023. PMID: 35927178 Free PMC article.
-
Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A.Science. 2006 May 5;312(5774):748-51. doi: 10.1126/science.1125162. Epub 2006 Apr 6. Science. 2006. PMID: 16601153
-
Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor.Nat Struct Mol Biol. 2008 Jan;15(1):109-11. doi: 10.1038/nsmb1326. Epub 2007 Dec 16. Nat Struct Mol Biol. 2008. PMID: 18084306 Free PMC article.
-
Tudor: a versatile family of histone methylation 'readers'.Trends Biochem Sci. 2013 Nov;38(11):546-55. doi: 10.1016/j.tibs.2013.08.002. Epub 2013 Sep 10. Trends Biochem Sci. 2013. PMID: 24035451 Free PMC article. Review.
-
Tudor-Containing Methyl-Lysine and Methyl-Arginine Reader Proteins: Disease Implications and Chemical Tool Development.ACS Chem Biol. 2025 Jan 17;20(1):33-47. doi: 10.1021/acschembio.4c00661. Epub 2024 Dec 24. ACS Chem Biol. 2025. PMID: 39718819 Review.
Cited by
-
Epigenetic control and cancer: the potential of histone demethylases as therapeutic targets.Pharmaceuticals (Basel). 2012 Sep 12;5(9):963-90. doi: 10.3390/ph5090963. Pharmaceuticals (Basel). 2012. PMID: 24280700 Free PMC article.
-
Unresolved Complexity in the Gene Regulatory Network Underlying EMT.Front Oncol. 2020 May 12;10:554. doi: 10.3389/fonc.2020.00554. eCollection 2020. Front Oncol. 2020. PMID: 32477926 Free PMC article. Review.
-
Understanding the Histone DNA Repair Code: H4K20me2 Makes Its Mark.Mol Cancer Res. 2018 Sep;16(9):1335-1345. doi: 10.1158/1541-7786.MCR-17-0688. Epub 2018 Jun 1. Mol Cancer Res. 2018. PMID: 29858375 Free PMC article. Review.
-
Computational Analysis and Predictive Cheminformatics Modeling of Small Molecule Inhibitors of Epigenetic Modifiers.PLoS One. 2016 Sep 13;11(9):e0083032. doi: 10.1371/journal.pone.0083032. eCollection 2016. PLoS One. 2016. PMID: 27622288 Free PMC article.
-
The COMPASS-like complex modulates fungal development and pathogenesis by regulating H3K4me3-mediated targeted gene expression in Magnaporthe oryzae.Mol Plant Pathol. 2021 Apr;22(4):422-439. doi: 10.1111/mpp.13035. Epub 2021 Feb 8. Mol Plant Pathol. 2021. PMID: 33559339 Free PMC article.
References
-
- Corsini L, Sattler M. Tudor hooks up with DNA repair. Nat Struct Mol Biol. 2007;14:98–99. - PubMed
-
- Torok MS, Grant PA. The generation and recognition of histone methylation. Results Probl Cell Differ. 2006;41:25–46. - PubMed
-
- Wiencke JK, Zheng S, Morrison Z, Yeh RF. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene. 2008;27:2412–2421. - PubMed
-
- Xavier de la C, Sergio L, Sara S-M, Marian AM-B. Do protein motifs read the histone code? BioEssays. 2005;27:164–175. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

