Kinetics of activated thrombin-activatable fibrinolysis inhibitor (TAFIa)-catalyzed cleavage of C-terminal lysine residues of fibrin degradation products and removal of plasminogen-binding sites
- PMID: 21467042
- PMCID: PMC3103306
- DOI: 10.1074/jbc.M110.215061
Kinetics of activated thrombin-activatable fibrinolysis inhibitor (TAFIa)-catalyzed cleavage of C-terminal lysine residues of fibrin degradation products and removal of plasminogen-binding sites
Abstract
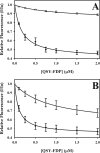
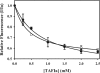
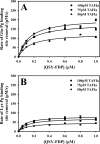
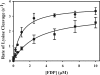
Partial digestion of fibrin by plasmin exposes C-terminal lysine residues, which comprise new binding sites for both plasminogen and tissue-type plasminogen activator (tPA). This binding increases the catalytic efficiency of plasminogen activation by 3000-fold compared with tPA alone. The activated thrombin-activatable fibrinolysis inhibitor (TAFIa) attenuates fibrinolysis by removing these residues, which causes a 97% reduction in tPA catalytic efficiency. The aim of this study was to determine the kinetics of TAFIa-catalyzed lysine cleavage from fibrin degradation products and the kinetics of loss of plasminogen-binding sites. We show that the k(cat) and K(m) of Glu(1)-plasminogen (Glu-Pg)-binding site removal are 2.34 s(-1) and 142.6 nm, respectively, implying a catalytic efficiency of 16.21 μm(-1) s(-1). The corresponding values of Lys(77)/Lys(78)-plasminogen (Lys-Pg)-binding site removal are 0.89 s(-1) and 96 nm implying a catalytic efficiency of 9.23 μm(-1) s(-1). These catalytic efficiencies of plasminogen-binding site removal by TAFIa are the highest of any TAFIa-catalyzed reaction with a biological substrate reported to date and suggest that plasmin-modified fibrin is a primary physiological substrate for TAFIa. We also show that the catalytic efficiency of cleavage of all C-terminal lysine residues, whether they are involved in plasminogen binding or not, is 1.10 μm(-1) s(-1). Interestingly, this value increases to 3.85 μm(-1) s(-1) in the presence of Glu-Pg. These changes are due to a decrease in K(m). This suggests that an interaction between TAFIa and plasminogen comprises a component of the reaction mechanism, the plausibility of which was established by showing that TAFIa binds both Glu-Pg and Lys-Pg.
Figures

Similar articles
-
Thrombin-activable fibrinolysis inhibitor zymogen does not play a significant role in the attenuation of fibrinolysis.J Biol Chem. 2008 Apr 4;283(14):8863-7. doi: 10.1074/jbc.M800127200. Epub 2008 Feb 5. J Biol Chem. 2008. PMID: 18252711 Free PMC article.
-
Activated thrombin-activatable fibrinolysis inhibitor (TAFIa) attenuates fibrin-dependent plasmin generation on thrombin-activated platelets.J Thromb Haemost. 2020 Sep;18(9):2364-2376. doi: 10.1111/jth.14950. J Thromb Haemost. 2020. PMID: 32506822 Free PMC article.
-
Activated thrombin-activatable fibrinolysis inhibitor reduces the ability of high molecular weight fibrin degradation products to protect plasmin from antiplasmin.J Biol Chem. 2004 Apr 2;279(14):13340-5. doi: 10.1074/jbc.M313211200. Epub 2004 Jan 8. J Biol Chem. 2004. PMID: 14715654
-
Insights into thrombin activatable fibrinolysis inhibitor function and regulation.J Thromb Haemost. 2013 Jun;11 Suppl 1:306-15. doi: 10.1111/jth.12216. J Thromb Haemost. 2013. PMID: 23809134 Review.
-
Modulation of fibrin cofactor activity in plasminogen activation.Ann N Y Acad Sci. 2001;936:247-60. doi: 10.1111/j.1749-6632.2001.tb03513.x. Ann N Y Acad Sci. 2001. PMID: 11460482 Review.
Cited by
-
Solulin increases clot stability in whole blood from humans and dogs with hemophilia.Blood. 2012 Apr 12;119(15):3622-8. doi: 10.1182/blood-2011-11-392308. Epub 2012 Jan 10. Blood. 2012. PMID: 22234684 Free PMC article. Clinical Trial.
-
Nutraceutical profile and evidence of alleviation of oxidative stress by Spirogyra porticalis (Muell.) Cleve inhabiting the high altitude Trans-Himalayan Region.Sci Rep. 2019 Mar 11;9(1):4091. doi: 10.1038/s41598-018-35595-x. Sci Rep. 2019. PMID: 30858387 Free PMC article.
-
Elevated cytokines, thrombin and PAI-1 in severe HCPS patients due to Sin Nombre virus.Viruses. 2015 Feb 10;7(2):559-89. doi: 10.3390/v7020559. Viruses. 2015. PMID: 25674766 Free PMC article.
-
Fibrinolytic Serine Proteases, Therapeutic Serpins and Inflammation: Fire Dancers and Firestorms.Front Cardiovasc Med. 2021 Mar 25;8:648947. doi: 10.3389/fcvm.2021.648947. eCollection 2021. Front Cardiovasc Med. 2021. PMID: 33869309 Free PMC article. Review.
-
Kinetic model facilitates analysis of fibrin generation and its modulation by clotting factors: implications for hemostasis-enhancing therapies.Mol Biosyst. 2014 Jul 29;10(9):2347-57. doi: 10.1039/c4mb00263f. Mol Biosyst. 2014. PMID: 24958246 Free PMC article.
References
-
- Collen D. (1999) Thromb. Haemost. 82, 259–270 - PubMed
-
- Fredenburgh J. C., Nesheim M. E. (1992) J. Biol. Chem. 267, 26150–26156 - PubMed
-
- Horrevoets A. J., Pannekoek H., Nesheim M. E. (1997) J. Biol. Chem. 272, 2176–2182 - PubMed
-
- Bajzar L., Manuel R., Nesheim M. E. (1995) J. Biol. Chem. 270, 14477–14484 - PubMed
-
- Mao S. S., Cooper C. M., Wood T., Shafer J. A., Gardell S. J. (1999) J. Biol. Chem. 274, 35046–35052 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

