Centromere identity, function, and epigenetic propagation across cell divisions
- PMID: 21467140
- PMCID: PMC3140419
- DOI: 10.1101/sqb.2010.75.038
Centromere identity, function, and epigenetic propagation across cell divisions
Abstract
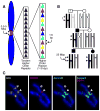
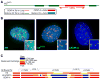
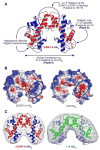
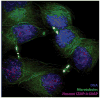
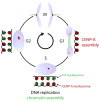
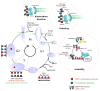
The key to understanding centromere identity is likely to lie in the chromatin containing the histone H3 variant CENP-A. CENP-A is the prime candidate to carry the epigenetic information that specifies the chromosomal location of the centromere in nearly all eukaryotic species, raising questions fundamental to understanding chromosome inheritance: How is the epigenetic centromere mark propagated? What physical properties of CENP-A-containing complexes are important for epigenetically marking centromeres? What are the molecules that recognize centromeric chromatin and serve as the foundation for the mitotic kinetochore? We discuss recent advances from our research groups that have yielded substantial insight into these questions and present our current understanding of the centromere. Future work promises an understanding of the molecular processes that confer fidelity to genome transmission at cell division.
Figures


References
-
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002b;9(6):1191–1200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
