mRNA isoform diversity can obscure detection of miRNA-mediated control of translation
- PMID: 21467217
- PMCID: PMC3096034
- DOI: 10.1261/rna.2567611
mRNA isoform diversity can obscure detection of miRNA-mediated control of translation
Abstract
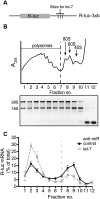
Reporter-based studies support inhibition of translation at the level of initiation as a substantial component of the miRNA mechanism, yet recent global analyses have suggested that they predominantly act through decreasing target mRNA stability. Cells commonly coexpress several processing isoforms of an mRNA, which may also differ in their regulatory untranslated regions (UTR). In particular, cancer cells are known to express high levels of short 3' UTR isoforms that evade miRNA-mediated regulation, whereas longer 3' UTRs predominate in nontransformed cells. To test whether mRNA isoform diversity can obscure detection of miRNA-mediated control at the level of translation, we assayed the responses of 11 endogenous let-7 targets to inactivation of this miRNA in HeLa cells, an intensively studied model system. We show that translational regulation in many cases appears to be modest when measuring the composite polysome profile of all extant isoforms of a given mRNA by density ultracentrifugation. In contrast, we saw clear effects at the level of translation initiation for multiple examples when selectively profiling mRNA isoforms carrying the 5' or 3' untranslated regions that were actually permissive to let-7 action, or when let-7 and a second targeting miRNA were jointly manipulated. Altogether, these results highlight a caveat to the mechanistic interpretation of data from global miRNA target analyses in transformed cells. Importantly, they reaffirm the importance of translational control as part of the miRNA mechanism in animal cells.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
