Directed culturing of microorganisms using metatranscriptomics
- PMID: 21467263
- PMCID: PMC3069634
- DOI: 10.1128/mBio.00012-11
Directed culturing of microorganisms using metatranscriptomics
Abstract
The vast majority of bacterial species remain uncultured, and this severely limits the investigation of their physiology, metabolic capabilities, and role in the environment. High-throughput sequencing of RNA transcripts (RNA-seq) allows the investigation of the diverse physiologies from uncultured microorganisms in their natural habitat. Here, we report the use of RNA-seq for characterizing the metatranscriptome of the simple gut microbiome from the medicinal leech Hirudo verbana and for utilizing this information to design a medium for cultivating members of the microbiome. Expression data suggested that a Rikenella-like bacterium, the most abundant but uncultured symbiont, forages on sulfated- and sialated-mucin glycans that are fermented, leading to the secretion of acetate. Histological stains were consistent with the presence of sulfated and sialated mucins along the crop epithelium. The second dominant symbiont, Aeromonas veronii, grows in two different microenvironments and is predicted to utilize either acetate or carbohydrates. Based on the metatranscriptome, a medium containing mucin was designed, which enabled the cultivation of the Rikenella-like bacterium. Metatranscriptomes shed light on microbial metabolism in situ and provide critical clues for directing the culturing of uncultured microorganisms. By choosing a condition under which the desired organism is rapidly proliferating and focusing on highly expressed genes encoding hydrolytic enzymes, binding proteins, and transporters, one can identify an organism's nutritional preferences and design a culture medium.
Importance: The number of prokaryotes on the planet has been estimated to exceed 10(30) cells, and the overwhelming majority of them have evaded cultivation, making it difficult to investigate their ecological, medical, and industrial relevance. The application of transcriptomics based on high-throughput sequencing of RNA transcripts (RNA-seq) to microorganisms in their natural environment can provide investigators with insight into their physiologies under optimal growth conditions. We utilized RNA-seq to learn more about the uncultured and cultured symbionts that comprise the relatively simple digestive-tract microbiome of the medicinal leech. The expression data revealed highly expressed hydrolytic enzymes and transporters that provided critical clues for the design of a culture medium enabling the isolation of the previously uncultured Rikenella-like symbiont. This directed culturing method will greatly aid efforts aimed at understanding uncultured microorganisms, including beneficial symbionts, pathogens, and ecologically relevant microorganisms, by facilitating genome sequencing, physiological characterization, and genetic manipulation of the previously uncultured microbes.
Figures
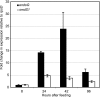




References
-
- Stingl U, Tripp HJ, Giovannoni SJ. 2007. Improvements of high-throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site. ISME J. 1:361–371 - PubMed
-
- Kaeberlein T, Lewis K, Epstein SS. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127–1129 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
