Transcriptional consequences of genomic structural aberrations in breast cancer
- PMID: 21467264
- PMCID: PMC3083084
- DOI: 10.1101/gr.113225.110
Transcriptional consequences of genomic structural aberrations in breast cancer
Abstract
Using a long-span, paired-end deep sequencing strategy, we have comprehensively identified cancer genome rearrangements in eight breast cancer genomes. Herein, we show that 40%-54% of these structural genomic rearrangements result in different forms of fusion transcripts and that 44% are potentially translated. We find that single segmental tandem duplication spanning several genes is a major source of the fusion gene transcripts in both cell lines and primary tumors involving adjacent genes placed in the reverse-order position by the duplication event. Certain other structural mutations, however, tend to attenuate gene expression. From these candidate gene fusions, we have found a fusion transcript (RPS6KB1-VMP1) recurrently expressed in ∼30% of breast cancers associated with potential clinical consequences. This gene fusion is caused by tandem duplication on 17q23 and appears to be an indicator of local genomic instability altering the expression of oncogenic components such as MIR21 and RPS6KB1.
Figures
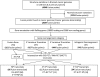
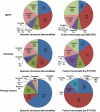
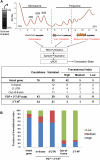
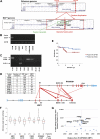
References
-
- Bärlund M, Forozan F, Kononen J, Bubendorf L, Chen Y, Bittner ML, Torhorst J, Haas P, Bucher C, Sauter G, et al. 2000. Detecting activation of ribosomal protein S6 kinase by complementary DNA and tissue microarray analysis. J Natl Cancer Inst 92: 1252–1259 - PubMed
-
- Bärlund M, Monni O, Weaver JD, Kauraniemi P, Sauter G, Heiskanen M, Kallioniemi OP, Kallioniemi A 2002. Cloning of BCAS3 (17q23) and BCAS4 (20q13) genes that undergo amplification, overexpression, and fusion in breast cancer. Genes Chromosomes Cancer 35: 311–317 - PubMed
-
- Beilharz TH, Preiss T 2004. Translational profiling: The genome-wide measure of the nascent proteome. Brief Funct Genomic Proteomic 3: 103–111 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
