Molecular chaperone Hsp104 can promote yeast prion generation
- PMID: 21467567
- PMCID: PMC3122315
- DOI: 10.1534/genetics.111.127779
Molecular chaperone Hsp104 can promote yeast prion generation
Abstract
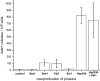
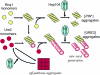
[URE3] is an amyloid-based prion of Ure2p, a regulator of nitrogen catabolism in Saccharomyces cerevisiae. The Ure2p of the human pathogen Candida albicans can also be a prion in S. cerevisiae. We find that overproduction of the disaggregating chaperone, Hsp104, increases the frequency of de novo [URE3] prion formation by the Ure2p of S. cerevisiae and that of C. albicans. This stimulation is strongly dependent on the presence of the [PIN(+)] prion, known from previous work to enhance [URE3] prion generation. Our data suggest that transient Hsp104 overproduction enhances prion generation through persistent effects on Rnq1 amyloid, as well as during overproduction by disassembly of amorphous Ure2 aggregates (generated during Ure2p overproduction), driving the aggregation toward the amyloid pathway. Overproduction of other major cytosolic chaperones of the Hsp70 and Hsp40 families (Ssa1p, Sse1p, and Ydj1p) inhibit prion formation, whereas another yeast Hsp40, Sis1p, modulates the effects of Hsp104p on both prion induction and prion curing in a prion-specific manner. The same factor may both enhance de novo prion generation and destabilize existing prion variants, suggesting that prion variants may be selected by changes in the chaperone network.
Figures


References
-
- Aguzzi A., Baumann F., Bremer J., 2008. The prion’s elusive reason for being. Annu. Rev. Neurosci. 31: 439–477 - PubMed
-
- Allen K. D., Chernova T. A., Tennant E. P., Wilkinson K. D., Chernoff Y. O., 2007. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J. Biol. Chem. 282: 3004–3013 - PubMed
-
- Arimon M., Grimminger V., Sanz F., Lashuel H. A., 2008. Hsp104 targets multiple intermediates on the amyloid pathway and suppresses the seeding capacity of Abeta fibrils and protofibrils. J. Mol. Biol. 384: 1157–1173 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

