GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-Chelatase and promote chlorophyll biosynthesis in Arabidopsis
- PMID: 21467578
- PMCID: PMC3101535
- DOI: 10.1105/tpc.110.082503
GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-Chelatase and promote chlorophyll biosynthesis in Arabidopsis
Abstract
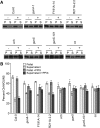
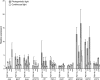
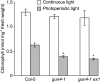
The GENOMES UNCOUPLED4 (GUN4) protein stimulates chlorophyll biosynthesis by activating Mg-chelatase, the enzyme that commits protoporphyrin IX to chlorophyll biosynthesis. This stimulation depends on GUN4 binding the ChlH subunit of Mg-chelatase and the porphyrin substrate and product of Mg-chelatase. After binding porphyrins, GUN4 associates more stably with chloroplast membranes and was proposed to promote interactions between ChlH and chloroplast membranes-the site of Mg-chelatase activity. GUN4 was also proposed to attenuate the production of reactive oxygen species (ROS) by binding and shielding light-exposed porphyrins from collisions with O₂. To test these proposals, we first engineered Arabidopsis thaliana plants that express only porphyrin binding-deficient forms of GUN4. Using these transgenic plants and particular mutants, we found that the porphyrin binding activity of GUN4 and Mg-chelatase contribute to the accumulation of chlorophyll, GUN4, and Mg-chelatase subunits. Also, we found that the porphyrin binding activity of GUN4 and Mg-chelatase affect the associations of GUN4 and ChlH with chloroplast membranes and have various effects on the expression of ROS-inducible genes. Based on our findings, we conclude that ChlH and GUN4 use distinct mechanisms to associate with chloroplast membranes and that mutant alleles of GUN4 and Mg-chelatase genes cause sensitivity to intense light by a mechanism that is potentially complex.
Figures

References
-
- Aarti D., Tanaka R., Ito H., Tanaka A. (2007). High light inhibits chlorophyll biosynthesis at the level of 5-aminolevulinate synthesis during de-etiolation in cucumber (Cucumis sativus) cotyledons. Photochem. Photobiol. 83: 171–176 - PubMed
-
- Aarti D.P., Tanaka R., Tanaka A. (2006). Effects of oxidative stress on chlorophyll biosynthesis in cucumber (Cucumis sativus) cotyledons. Physiol. Plant. 128: 186–197
-
- Alawady A.E., Grimm B. (2005). Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. Plant J. 41: 282–290 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

