Unlocking the barley genome by chromosomal and comparative genomics
- PMID: 21467582
- PMCID: PMC3101540
- DOI: 10.1105/tpc.110.082537
Unlocking the barley genome by chromosomal and comparative genomics
Abstract
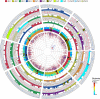
We used a novel approach that incorporated chromosome sorting, next-generation sequencing, array hybridization, and systematic exploitation of conserved synteny with model grasses to assign ~86% of the estimated ~32,000 barley (Hordeum vulgare) genes to individual chromosome arms. Using a series of bioinformatically constructed genome zippers that integrate gene indices of rice (Oryza sativa), sorghum (Sorghum bicolor), and Brachypodium distachyon in a conserved synteny model, we were able to assemble 21,766 barley genes in a putative linear order. We show that the barley (H) genome displays a mosaic of structural similarity to hexaploid bread wheat (Triticum aestivum) A, B, and D subgenomes and that orthologous genes in different grasses exhibit signatures of positive selection in different lineages. We present an ordered, information-rich scaffold of the barley genome that provides a valuable and robust framework for the development of novel strategies in cereal breeding.
Figures


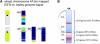
References
-
- Abrouk M., Murat F., Pont C., Messing J., Jackson S., Faraut T., Tannier E., Plomion C., Cooke R., Feuillet C., Salse J. (2010). Palaeogenomics of plants: Synteny-based modelling of extinct ancestors. Trends Plant Sci. 15: 479–487 - PubMed
-
- Agalou A., et al. (2008). A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 66: 87–103 - PubMed
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 - PubMed
-
- Beales J., Turner A., Griffiths S., Snape J.W., Laurie D.A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115: 721–733 - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

