Engineering scalable biological systems
- PMID: 21468204
- PMCID: PMC3056087
- DOI: 10.4161/bbug.1.6.13086
Engineering scalable biological systems
Abstract
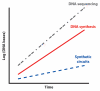
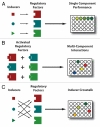
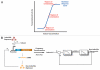
Synthetic biology is focused on engineering biological organisms to study natural systems and to provide new solutions for pressing medical, industrial and environmental problems. At the core of engineered organisms are synthetic biological circuits that execute the tasks of sensing inputs, processing logic and performing output functions. In the last decade, significant progress has been made in developing basic designs for a wide range of biological circuits in bacteria, yeast and mammalian systems. However, significant challenges in the construction, probing, modulation and debugging of synthetic biological systems must be addressed in order to achieve scalable higher-complexity biological circuits. Furthermore, concomitant efforts to evaluate the safety and biocontainment of engineered organisms and address public and regulatory concerns will be necessary to ensure that technological advances are translated into real-world solutions.
Keywords: biological circuits; biological modulators; biological probes; engineered organisms; high-throughput design; modelling; regulatory issues; synthetic biology.
© 2010 Landes Bioscience
Figures
Comment on
-
Next-generation synthetic gene networks.Nat Biotechnol. 2009 Dec;27(12):1139-50. doi: 10.1038/nbt.1591. Nat Biotechnol. 2009. PMID: 20010597 Free PMC article.
Similar articles
-
Synthetic circuits, devices and modules.Protein Cell. 2010 Nov;1(11):974-8. doi: 10.1007/s13238-010-0133-8. Epub 2010 Dec 10. Protein Cell. 2010. PMID: 21153514 Free PMC article. Review.
-
Synthetic gene networks in mammalian cells.Curr Opin Biotechnol. 2010 Oct;21(5):690-6. doi: 10.1016/j.copbio.2010.07.006. Epub 2010 Aug 4. Curr Opin Biotechnol. 2010. PMID: 20691580 Review.
-
Designing cell function: assembly of synthetic gene circuits for cell biology applications.Nat Rev Mol Cell Biol. 2018 Aug;19(8):507-525. doi: 10.1038/s41580-018-0024-z. Nat Rev Mol Cell Biol. 2018. PMID: 29858606 Review.
-
Programming gene expression in multicellular organisms for physiology modulation through engineered bacteria.Nat Commun. 2021 May 11;12(1):2689. doi: 10.1038/s41467-021-22894-7. Nat Commun. 2021. PMID: 33976154 Free PMC article.
-
Multiplexed Gene Expression Tuning with Orthogonal Synthetic Gene Circuits.ACS Synth Biol. 2020 Apr 17;9(4):930-939. doi: 10.1021/acssynbio.9b00534. Epub 2020 Mar 23. ACS Synth Biol. 2020. PMID: 32167761 Free PMC article.
Cited by
-
Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology.Nucleic Acids Res. 2013 Mar 1;41(5):3471-81. doi: 10.1093/nar/gkt052. Epub 2013 Jan 31. Nucleic Acids Res. 2013. PMID: 23371936 Free PMC article.
-
Rational diversification of a promoter providing fine-tuned expression and orthogonal regulation for synthetic biology.PLoS One. 2012;7(3):e33279. doi: 10.1371/journal.pone.0033279. Epub 2012 Mar 19. PLoS One. 2012. PMID: 22442681 Free PMC article.
-
Diagnostics for stochastic genome-scale modeling via model slicing and debugging.PLoS One. 2014 Nov 4;9(11):e110380. doi: 10.1371/journal.pone.0110380. eCollection 2014. PLoS One. 2014. PMID: 25368989 Free PMC article.
References
-
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–778. - PubMed
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
-
- Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, et al. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260:500–507. - PubMed
-
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
