Promoter architectures in the yeast ribosomal expression program
- PMID: 21468232
- PMCID: PMC3062397
- DOI: 10.4161/trns.2.2.14486
Promoter architectures in the yeast ribosomal expression program
Abstract
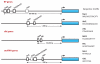
Ribosome biogenesis begins with the orchestrated expression of hundreds of genes, including the three large classes of ribosomal protein, ribosome biogenesis and snoRNA genes. Current knowledge about the corresponding promoters suggests the existence of novel class-specific transcriptional strategies and crosstalk between telomere length and cell growth control.
Figures

References
-
- Rudra D, Warner JR. What better measure than ribosome synthesis? Genes Dev. 2004;18:2431–2436. - PubMed
-
- Lascaris RF, Mager WH, Planta RJ. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics. 1999;15:267–277. - PubMed
-
- Lieb JD, Liu X, Botstein D, Brown PO. Promoterspecific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
