Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly
- PMID: 21468301
- PMCID: PMC3066130
- DOI: 10.1371/journal.pbio.1000603
Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly
Abstract
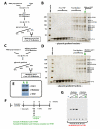
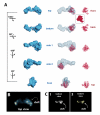
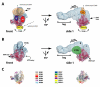
The macromolecular assembly required to initiate transcription of protein-coding genes, known as the Pre-Initiation Complex (PIC), consists of multiple protein complexes and is approximately 3.5 MDa in size. At the heart of this assembly is the Mediator complex, which helps regulate PIC activity and interacts with the RNA polymerase II (pol II) enzyme. The structure of the human Mediator-pol II interface is not well-characterized, whereas attempts to structurally define the Mediator-pol II interaction in yeast have relied on incomplete assemblies of Mediator and/or pol II and have yielded inconsistent interpretations. We have assembled the complete, 1.9 MDa human Mediator-pol II-TFIIF complex from purified components and have characterized its structural organization using cryo-electron microscopy and single-particle reconstruction techniques. The orientation of pol II within this assembly was determined by crystal structure docking and further validated with projection matching experiments, allowing the structural organization of the entire human PIC to be envisioned. Significantly, pol II orientation within the Mediator-pol II-TFIIF assembly can be reconciled with past studies that determined the location of other PIC components relative to pol II itself. Pol II surfaces required for interacting with TFIIB, TFIIE, and promoter DNA (i.e., the pol II cleft) are exposed within the Mediator-pol II-TFIIF structure; RNA exit is unhindered along the RPB4/7 subunits; upstream and downstream DNA is accessible for binding additional factors; and no major structural re-organization is necessary to accommodate the large, multi-subunit TFIIH or TFIID complexes. The data also reveal how pol II binding excludes Mediator-CDK8 subcomplex interactions and provide a structural basis for Mediator-dependent control of PIC assembly and function. Finally, parallel structural analysis of Mediator-pol II complexes lacking TFIIF reveal that TFIIF plays a key role in stabilizing pol II orientation within the assembly.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

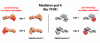

References
-
- Gilmour D. S. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. - PubMed
-
- Baek H. J, Kang Y. K, Roeder R. G. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem. 2006;281:15172–15181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

