Proximity ligation in situ assay for monitoring the global DNA methylation in cells
- PMID: 21470401
- PMCID: PMC3080290
- DOI: 10.1186/1472-6750-11-31
Proximity ligation in situ assay for monitoring the global DNA methylation in cells
Abstract
Background: DNA methylation has a central role in the epigenetic control of mammalian gene expression, and is required for X inactivation, genomics imprinting and silencing of retrotransposons and repetitive sequences. Thus, several technologies have been developed to measure the degree of DNA methylation.
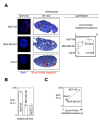
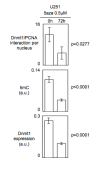
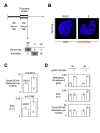
Results: We here present the development of the detection of protein-protein interactions via the adaptation of the proximity ligation in situ technology to evaluate the DNA methylation status in cells since the quantification of Dnmt1/PCNA interaction in cells reflects the degree of DNA methylation.
Conclusion: This method being directly realizable on cells, it appears that it could suggest a wide range of applications in basic research and drug development. More particularly, this method is specially adapted for the investigations realized from a weak quantity of biologic materiel such as stem cells or primary cultured tumor cells for examples.
Figures


Similar articles
-
Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements.Mol Cell Biol. 2002 Jan;22(2):480-91. doi: 10.1128/MCB.22.2.480-491.2002. Mol Cell Biol. 2002. PMID: 11756544 Free PMC article.
-
DNA methylation inhibitor 5-Aza-2'-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B.Mol Cell Biol. 2008 Jan;28(2):752-71. doi: 10.1128/MCB.01799-07. Epub 2007 Nov 8. Mol Cell Biol. 2008. PMID: 17991895 Free PMC article.
-
Induction of the estrogen receptor by ablation of DNMT1 in ER-negative breast cancer cells.Cancer Biol Ther. 2003 Sep-Oct;2(5):557-8. doi: 10.4161/cbt.2.5.588. Cancer Biol Ther. 2003. PMID: 14614326 No abstract available.
-
DNA methyltransferase-1 inhibitors as epigenetic therapy for cancer.Curr Cancer Drug Targets. 2013 May;13(4):379-99. doi: 10.2174/15680096113139990077. Curr Cancer Drug Targets. 2013. PMID: 23517596 Review.
-
An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function.Epigenetics. 2012 Sep;7(9):994-1007. doi: 10.4161/epi.21568. Epub 2012 Aug 16. Epigenetics. 2012. PMID: 22894906 Free PMC article. Review.
Cited by
-
The interplay between the lysine demethylase KDM1A and DNA methyltransferases in cancer cells is cell cycle dependent.Oncotarget. 2016 Sep 13;7(37):58939-58952. doi: 10.18632/oncotarget.10624. Oncotarget. 2016. PMID: 27449289 Free PMC article.
-
Epigenetic regulation of estrogen signaling in breast cancer.Epigenetics. 2013 Mar;8(3):237-45. doi: 10.4161/epi.23790. Epub 2013 Jan 30. Epigenetics. 2013. PMID: 23364277 Free PMC article. Review.
-
The autophagy GABARAPL1 gene is epigenetically regulated in breast cancer models.BMC Cancer. 2015 Oct 17;15:729. doi: 10.1186/s12885-015-1761-4. BMC Cancer. 2015. PMID: 26474850 Free PMC article.
-
Identification of TET1 Partners That Control Its DNA-Demethylating Function.Genes Cancer. 2013 May;4(5-6):235-41. doi: 10.1177/1947601913489020. Genes Cancer. 2013. PMID: 24069510 Free PMC article.
-
Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma.Clin Epigenetics. 2018 Feb 9;10:17. doi: 10.1186/s13148-018-0450-y. eCollection 2018. Clin Epigenetics. 2018. PMID: 29449903 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

