Genomic analysis of highly virulent Georgia 2007/1 isolate of African swine fever virus
- PMID: 21470447
- PMCID: PMC3379899
- DOI: 10.3201/eid1704.101283
Genomic analysis of highly virulent Georgia 2007/1 isolate of African swine fever virus
Abstract
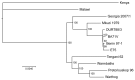
African swine fever is widespread in Africa but has occasionally been introduced into other continents. In June 2007, African swine fever was isolated in the Caucasus Region of the Republic of Georgia and subsequently in neighboring countries (Armenia, Azerbaijan, and 9 states of the Russian Federation). Previous data for sequencing of 3 genes indicated that the Georgia 2007/1 isolate is closely related to isolates of genotype II, which has been identified in Mozambique, Madagascar, and Zambia. We report the complete genomic coding sequence of the Georgia 2007/1 isolate and comparison with other isolates. A genome sequence of 189,344 bp encoding 166 open reading frames (ORFs) was obtained. Phylogeny based on concatenated sequences of 125 conserved ORFs showed that this isolate clustered most closely with the Mkuzi 1979 isolate. Some ORFs clustered differently, suggesting that recombination may have occurred. Results provide a baseline for monitoring genomic changes in this virus.
Figures

References
-
- Dixon LK, Escribano JM, Martins C, Rock DL, Salas ML, Wilkinson PJ. Asfarviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. London: Elsevier/Academic Press; 2005. p. 135–43.
-
- ProMED-mail. African swine fever—Russia (05): (KB), conf., culled. 2010. Apr 6 [cited 2011 Jan 6]. http://www.promedmail.org, archive no. 20100406.1107.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
