Utilizing natural diversity to evolve protein function: applications towards thermostability
- PMID: 21470898
- PMCID: PMC3173975
- DOI: 10.1016/j.cbpa.2011.03.005
Utilizing natural diversity to evolve protein function: applications towards thermostability
Abstract
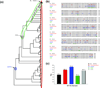
Protein evolution relies on designing a library of sequences that capture meaningful functional diversity in a limited number of protein variants. Several approaches take advantage of the sequence space already explored through natural selection by incorporating sequence diversity available from modern genomes (and their ancestors) when designing these libraries. The success of these approaches is, partly, owing to the fact that modern sequence diversity has already been subjected to evolutionary selective forces and thus the diversity has already been deemed 'fit to survive'. Five of these approaches will be discussed in this review to highlight how protein engineers can use evolutionary sequence history/diversity of homologous proteins in unique ways to design protein libraries.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- Lutz S, Bornscheuer U. The Protein Engineering Handbook. Weinheim: Wiley-VCH; 2009.
-
- Szostak JW. Functional information: molecular messages. Nature. 2003;423:689. - PubMed
-
- Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. - PubMed
-
- Ness JE, Kim S, Gottman A, Pak R, Krebber A, Borchert TV, Govindarajan S, Mundorff EC, Minshull J. Synthetic shuffling expands functional protein diversity by allowing amino acids to recombine independently. Nat Biotechnol. 2002;20:1251–1255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

