Intercompartmental recombination of HIV-1 contributes to env intrahost diversity and modulates viral tropism and sensitivity to entry inhibitors
- PMID: 21471230
- PMCID: PMC3126287
- DOI: 10.1128/JVI.00131-11
Intercompartmental recombination of HIV-1 contributes to env intrahost diversity and modulates viral tropism and sensitivity to entry inhibitors
Abstract
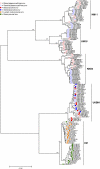
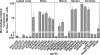
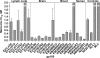
HIV-1 circulates within an infected host as a genetically heterogeneous viral population. Viral intrahost diversity is shaped by substitutional evolution and recombination. Although many studies have speculated that recombination could have a significant impact on viral phenotype, this has never been definitively demonstrated. We report here phylogenetic and subsequent phenotypic analyses of envelope genes obtained from HIV-1 populations present in different anatomical compartments. Assessment of env compartmentalization from immunologically discrete tissues was assessed utilizing a single genome amplification approach, minimizing in vitro-generated artifacts. Genetic compartmentalization of variants was frequently observed. In addition, multiple incidences of intercompartment recombination, presumably facilitated by low-level migration of virus or infected cells between different anatomic sites and coinfection of susceptible cells by genetically divergent strains, were identified. These analyses demonstrate that intercompartment recombination is a fundamental evolutionary mechanism that helps to shape HIV-1 env intrahost diversity in natural infection. Analysis of the phenotypic consequences of these recombination events showed that genetic compartmentalization often correlates with phenotypic compartmentalization and that intercompartment recombination results in phenotype modulation. This represents definitive proof that recombination can generate novel combinations of phenotypic traits which differ subtly from those of parental strains, an important phenomenon that may have an impact on antiviral therapy and contribute to HIV-1 persistence in vivo.
Figures

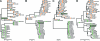


References
-
- Abbate I., et al. 2005. Cell membrane proteins and quasispecies compartmentalization of CSF and plasma HIV-1 from aids patients with neurological disorders. Infect. Genet. Evol. 5:247–253 - PubMed
-
- Ball J. K., Holmes E. C., Whitwell H., Desselberger U. 1994. Genomic variation of human immunodeficiency virus type 1 (HIV-1): molecular analyses of HIV-1 in sequential blood samples and various organs obtained at autopsy. J. Gen. Virol. 75(Pt. 4):67–79 - PubMed
-
- Bart J., et al. 2002. An oncological view on the blood-testis barrier. Lancet Oncol. 3:357–363 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

