Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea
- PMID: 21472016
- PMCID: PMC3176514
- DOI: 10.1038/ismej.2011.41
Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea
Abstract
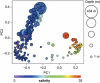
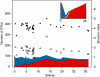
Salinity is a major factor controlling the distribution of biota in aquatic systems, and most aquatic multicellular organisms are either adapted to life in saltwater or freshwater conditions. Consequently, the saltwater-freshwater mixing zones in coastal or estuarine areas are characterized by limited faunal and floral diversity. Although changes in diversity and decline in species richness in brackish waters is well documented in aquatic ecology, it is unknown to what extent this applies to bacterial communities. Here, we report a first detailed bacterial inventory from vertical profiles of 60 sampling stations distributed along the salinity gradient of the Baltic Sea, one of world's largest brackish water environments, generated using 454 pyrosequencing of partial (400 bp) 16S rRNA genes. Within the salinity gradient, bacterial community composition altered at broad and finer-scale phylogenetic levels. Analogous to faunal communities within brackish conditions, we identified a bacterial brackish water community comprising a diverse combination of freshwater and marine groups, along with populations unique to this environment. As water residence times in the Baltic Sea exceed 3 years, the observed bacterial community cannot be the result of mixing of fresh water and saltwater, but our study represents the first detailed description of an autochthonous brackish microbiome. In contrast to the decline in the diversity of multicellular organisms, reduced bacterial diversity at brackish conditions could not be established. It is possible that the rapid adaptation rate of bacteria has enabled a variety of lineages to fill what for higher organisms remains a challenging and relatively unoccupied ecological niche.
Figures


References
-
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. - PubMed
-
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2009;4:171–181. - PubMed
-
- Arnds J, Knittel K, Buck U, Winkel M, Amann R. Development of a 16S rRNA-targeted probe set for Verrucomicrobia and its application for fluorescence in situ hybridization in a humic lake. Syst Appl Microbiol. 2010;33:139–148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

