Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms
- PMID: 21474065
- PMCID: PMC3175429
- DOI: 10.1016/j.molcel.2011.03.004
Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms
Abstract
In contrast to the active conformations of protein kinases, which are essentially the same for all kinases, inactive kinase conformations are structurally diverse. Some inactive conformations are, however, observed repeatedly in different kinases, perhaps reflecting an important role in catalysis. In this review, we analyze one of these recurring conformations, first identified in CDK and Src kinases, which turned out to be central to understanding of how kinase domain of the EGF receptor is activated. This mechanism, which involves the stabilization of the active conformation of an α helix, has features in common with mechanisms operative in several other kinases.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

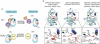

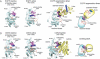


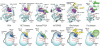
References
-
- Aifa S, Aydin J, Nordvall G, Lundstrom I, Svensson SP, Hermanson O. A basic peptide within the juxtamembrane region is required for EGF receptor dimerization. Exp Cell Res. 2005;302:108–114. - PubMed
-
- Bargmann CI, Hung MC, Weinberg RA. Multiple independent activations of the neu oncogene by a point mutation altering the transmembrane domain of p185. Cell. 1986a;45:649–657. - PubMed
-
- Bargmann CI, Hung MC, Weinberg RA. The neu oncogene encodes an epidermal growth factor receptor-related protein. Nature. 1986b;319:226–230. - PubMed
-
- Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–862. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

