Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size
- PMID: 21474067
- PMCID: PMC3750737
- DOI: 10.1016/j.molcel.2011.03.017
Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size
Abstract
Mammalian target of rapamycin (mTOR) is a serine/threonine kinase that exists in two separate complexes, mTORC1 and mTORC2, that function to control cell size and growth in response to growth factors, nutrients, and cellular energy levels. Low molecular weight GTP-binding proteins of the Rheb and Rag families are key regulators of the mTORC1 complex, but regulation of mTORC2 is poorly understood. Here, we report that Rac1, a member of the Rho family of GTPases, is a critical regulator of both mTORC1 and mTORC2 in response to growth-factor stimulation. Deletion of Rac1 in primary cells using an inducible-Cre/Lox approach inhibits basal and growth-factor activation of both mTORC1 and mTORC2. Rac1 appears to bind directly to mTOR and to mediate mTORC1 and mTORC2 localization at specific membranes. Binding of Rac1 to mTOR does not depend on the GTP-bound state of Rac1, but on the integrity of its C-terminal domain. This function of Rac1 provides a means to regulate mTORC1 and mTORC2 simultaneously.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
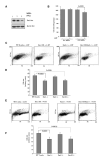


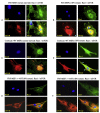



Comment in
-
Cell growth: RAC1 sizes up mTOR.Nat Rev Mol Cell Biol. 2011 Jun;12(6):343. doi: 10.1038/nrm3124. Epub 2011 May 18. Nat Rev Mol Cell Biol. 2011. PMID: 21587295 No abstract available.
References
-
- Avruch J, Hara K, Lin Y, Liu M, Long X, Ortiz-Vega S, Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. - PubMed
-
- Benvenuti F, Hugues S, Walmsley M, Ruf S, Fetler L, Popoff M, Tybulewicz VL, Amigorena S. Requirement of Rac1 and Rac2 expression by mature dendritic cells for T cell priming. Science. 2004;305:1150–1153. - PubMed
-
- Berven LA, Willard FS, Crouch MF. Role of the p70(S6K) pathway in regulating the actin cytoskeleton and cell migration. Exp. Cell Res. 2004;296:183–195. - PubMed
-
- Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

