Complete genome sequence of Leadbetterella byssophila type strain (4M15)
- PMID: 21475582
- PMCID: PMC3072089
- DOI: 10.4056/sigs.1413518
Complete genome sequence of Leadbetterella byssophila type strain (4M15)
Abstract
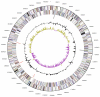
Leadbetterella byssophila Weon et al. 2005 is the type species of the genus Leadbetterella of the family Cytophagaceae in the phylum Bacteroidetes. Members of the phylum Bacteroidetes are widely distributed in nature, especially in aquatic environments. They are of special interest for their ability to degrade complex biopolymers. L. byssophila occupies a rather isolated position in the tree of life and is characterized by its ability to hydrolyze starch and gelatine, but not agar, cellulose or chitin. Here we describe the features of this organism, together with the complete genome sequence, and annotation. L. byssophila is already the 16(th) member of the family Cytophagaceae whose genome has been sequenced. The 4,059,653 bp long single replicon genome with its 3,613 protein-coding and 53 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: Cytophagaceae; GEBA; Gram-negative; aerobic; flexirubin; mesophile; non-motile; non-sporulating.
Figures


References
-
- Ludwig W, Klenk HP. Overview: a phylogenetic backbone and taxonomic framework for procaryotic systematics. In: Bergey's Manual of Systematic Bacteriology 2001. Boone DR, Castenholz RW, Garrity GM (eds), 2nd edn, vol. 1, pp. 49-65. New York: Springer.
-
- Garrity GM, Lilburn TG, Cole JR, Harrison SH, Euzéby J, Tindall BJ. Taxonomic outline of the Bacteria and Archaea, Release 7.7 March 6, 2007. Part 11 - The Bacteria: Phyla 'Planctomycetes', 'Chlamydiae', 'Spirochaetes', 'Fibrobacteres', 'Acidobacteria', 'Bacteroidetes', 'Fusobacteria', 'Verrucomicrobia', 'Dictyoglomi', 'Gemmatimonadetes', and 'Lentisphaerae'. http://www.taxonomicoutline.org/index.php/toba/article/viewFile/188/220 2007.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

