Molecular motors: directing traffic during RNA localization
- PMID: 21476929
- PMCID: PMC3181154
- DOI: 10.3109/10409238.2011.572861
Molecular motors: directing traffic during RNA localization
Abstract
RNA localization, the enrichment of RNA in a specific subcellular region, is a mechanism for the establishment and maintenance of cellular polarity in a variety of systems. Ultimately, this results in a universal method for spatially restricting gene expression. Although the consequences of RNA localization are well-appreciated, many of the mechanisms that are responsible for carrying out polarized transport remain elusive. Several recent studies have illuminated the roles that molecular motor proteins play in the process of RNA localization. These studies have revealed complex mechanisms in which the coordinated action of one or more motor proteins can act at different points in the localization process to direct RNAs to their final destination. In this review, we discuss recent findings from several different systems in an effort to clarify pathways and mechanisms that control the directed movement of RNA.
Conflict of interest statement
The authors report that they have no conflicts of interest. Our work on RNA localization is funded by a grant from the National Institutes of Health (GM071049) to K.L.M. J.A.G. was a predoctoral trainee supported in part by grant T32-GM07601.
Figures

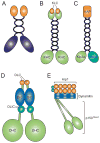
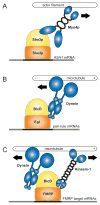
References
-
- Adelstein RS, Eisenberg E. Regulation and kinetics of the actin-myosin-ATP interaction. Annu Rev Biochem. 1980;49:921–956. - PubMed
-
- Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J Cell Sci. 2002;115:3817–3827. - PubMed
-
- Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Annu Rev Biochem. 1998;67:335–394. - PubMed
-
- Berezuk MA, Schroer TA. Dynactin enhances the processivity of kinesin-2. Traffic. 2007;8:124–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
