Genetic composition of contemporary swine influenza viruses in the West Central region of the United States of America
- PMID: 21477138
- PMCID: PMC3079267
- DOI: 10.1111/j.1750-2659.2010.00189.x
Genetic composition of contemporary swine influenza viruses in the West Central region of the United States of America
Abstract
Background: Because of continuous circulation in different animal species and humans, influenza viruses have host-specific phenotypic and genetic features. Reassortment of the genome segments can significantly change virus phenotype, potentially generating virus with pandemic potential. In 2009, a new pandemic influenza virus emerged.
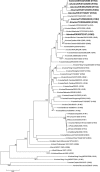
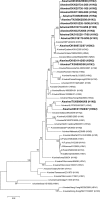
Objectives: In this study, we attempted to find precursor viruses or genes of pandemic H1N1 influenza 2009 among 25 swine influenza viruses, isolated in the West Central region of the United States of America (USA), between 2007 and 2009. The Phylogenetically Similar Triple-Reassortant Internal Genes (PSTRIG) cassette of all the viruses studied here as well as the PSTRIG cassette of pandemic H1N1 viruses have close but equidistant phylogenetic relationships to the early triple-reassortant swine H3N2 influenza A isolated in the USA in 1998.
Methods: Samples (nasal swabs and lung tissue lavage) were taken from swine with or without clinical signs of respiratory disease via farmer-funded syndromic surveillance. All studied viruses were isolated in Madin-Darby Canine Kidney cell cultures from the above-mentioned samples according to standard protocols recommended for influenza virus isolation. Sequences were obtained using BigDye Terminator v3.1 Cycle Sequencing kit. Phylogenetic trees were built with MEGA 4.0 software using maximum composite likelihood algorithm and neighbor-joining method for tree topology reconstruction.
Results: Among the 25 viruses studied, we have not found any gene segments of Eurasian origin. Our results suggest that pandemic H1N1 viruses diverged and are not directly descended from swine viruses that have been circulating in USA since 1998.
© 2011 Blackwell Publishing Ltd.
Figures



References
-
- Vincent AL, Ma W, Lager M, Janke BH, Richt JA. Swine influenza viruses a North American perspective. Adv Virus Res 2008; 72:127–154. Review. - PubMed
-
- Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol 2000; 74:29–46. - PubMed
-
- Smith GJ, Vijaykrishna D, Bahl J et al. Origins and evolutionary genomics of the 2009 swine‐origin H1N1 influenza A epidemic. Nature 2009; 459:1122–1125. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

