Identification of staphylococcal nuclease domain-containing 1 (SND1) as a Metadherin-interacting protein with metastasis-promoting functions
- PMID: 21478147
- PMCID: PMC3103372
- DOI: 10.1074/jbc.M111.240077
Identification of staphylococcal nuclease domain-containing 1 (SND1) as a Metadherin-interacting protein with metastasis-promoting functions
Abstract
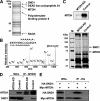
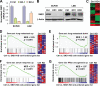
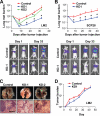
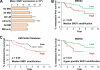
Metastasis is the deadliest and most poorly understood feature of malignant diseases. Recent work has shown that Metadherin (MTDH) is overexpressed in over 40% of breast cancer patients and promotes metastasis and chemoresistance in experimental models of breast cancer progression. Here we applied mass spectrometry-based screen to identify staphylococcal nuclease domain-containing 1 (SND1) as a candidate MTDH-interacting protein. After confirming the interaction between SND1 and MTDH, we tested the role of SND1 in breast cancer and found that it strongly promotes lung metastasis. SND1 was further shown to promote resistance to apoptosis and to regulate the expression of genes associated with metastasis and chemoresistance. Analyses of breast cancer clinical microarray data indicated that high expression of SND1 in primary tumors is strongly associated with reduced metastasis-free survival in multiple large scale data sets. Thus, we have uncovered SND1 as a novel MTDH-interacting protein and shown that it is a functionally and clinically significant mediator of metastasis.
Figures


References
-
- Jemal A., Siegel R., Xu J., Ward E. (2010) CA Cancer J. Clin. 60, 277–300 - PubMed
-
- Nguyen D. X., Bos P. D., Massagué J. (2009) Nat. Rev. Cancer 9, 274–284 - PubMed
-
- Brown D. M., Ruoslahti E. (2004) Cancer Cell 5, 365–374 - PubMed
-
- Kang D. C., Su Z. Z., Sarkar D., Emdad L., Volsky D. J., Fisher P. B. (2005) Gene 353, 8–15 - PubMed
-
- Britt D. E., Yang D. F., Yang D. Q., Flanagan D., Callanan H., Lim Y. P., Lin S. H., Hixson D. C. (2004) Exp. Cell Res. 300, 134–148 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

