Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
- PMID: 21478274
- PMCID: PMC3084024
- DOI: 10.1101/gad.2045111
Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Abstract
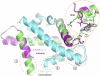
In higher eukaryotes, the centromere is epigenetically specified by the histone H3 variant Centromere Protein-A (CENP-A). Deposition of CENP-A to the centromere requires histone chaperone HJURP (Holliday junction recognition protein). The crystal structure of an HJURP-CENP-A-histone H4 complex shows that HJURP binds a CENP-A-H4 heterodimer. The C-terminal β-sheet domain of HJURP caps the DNA-binding region of the histone heterodimer, preventing it from spontaneous association with DNA. Our analysis also revealed a novel site in CENP-A that distinguishes it from histone H3 in its ability to bind HJURP. These findings provide key information for specific recognition of CENP-A and mechanistic insights into the process of centromeric chromatin assembly.
Figures
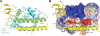
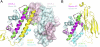

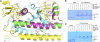
Similar articles
-
Putting CENP-A in its place.Cell Mol Life Sci. 2013 Feb;70(3):387-406. doi: 10.1007/s00018-012-1048-8. Epub 2012 Jun 23. Cell Mol Life Sci. 2013. PMID: 22729156 Free PMC article. Review.
-
HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres.Proc Natl Acad Sci U S A. 2010 Jan 26;107(4):1349-54. doi: 10.1073/pnas.0913709107. Epub 2010 Jan 6. Proc Natl Acad Sci U S A. 2010. PMID: 20080577 Free PMC article.
-
Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition.EMBO J. 2013 Jul 31;32(15):2113-24. doi: 10.1038/emboj.2013.142. Epub 2013 Jun 14. EMBO J. 2013. PMID: 23771058 Free PMC article.
-
HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly.Dev Cell. 2012 Apr 17;22(4):749-62. doi: 10.1016/j.devcel.2012.02.001. Epub 2012 Mar 8. Dev Cell. 2012. PMID: 22406139 Free PMC article.
-
Structure of the CENP-A nucleosome and its implications for centromeric chromatin architecture.Genes Genet Syst. 2011;86(6):357-64. doi: 10.1266/ggs.86.357. Genes Genet Syst. 2011. PMID: 22451475 Review.
Cited by
-
Putting CENP-A in its place.Cell Mol Life Sci. 2013 Feb;70(3):387-406. doi: 10.1007/s00018-012-1048-8. Epub 2012 Jun 23. Cell Mol Life Sci. 2013. PMID: 22729156 Free PMC article. Review.
-
Chromatin assembly: Journey to the CENter of the chromosome.J Cell Biol. 2016 Jul 4;214(1):13-24. doi: 10.1083/jcb.201605005. J Cell Biol. 2016. PMID: 27377247 Free PMC article. Review.
-
How two become one: HJURP dimerization drives CENP-A assembly.EMBO J. 2013 Jul 31;32(15):2090-2. doi: 10.1038/emboj.2013.150. Epub 2013 Jun 21. EMBO J. 2013. PMID: 23792427 Free PMC article.
-
Histone supply: Multitiered regulation ensures chromatin dynamics throughout the cell cycle.J Cell Biol. 2019 Jan 7;218(1):39-54. doi: 10.1083/jcb.201807179. Epub 2018 Sep 26. J Cell Biol. 2019. PMID: 30257851 Free PMC article. Review.
-
Identification of an ubinuclein 1 region required for stability and function of the human HIRA/UBN1/CABIN1/ASF1a histone H3.3 chaperone complex.Biochemistry. 2012 Mar 27;51(12):2366-77. doi: 10.1021/bi300050b. Epub 2012 Mar 16. Biochemistry. 2012. PMID: 22401310 Free PMC article.
References
-
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL Jr, Cleveland DW 2004. Structural determinants for generating centromeric chromatin. Nature 430: 578–582 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
