A system for the continuous directed evolution of biomolecules
- PMID: 21478873
- PMCID: PMC3084352
- DOI: 10.1038/nature09929
A system for the continuous directed evolution of biomolecules
Abstract
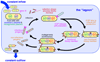
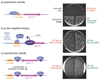
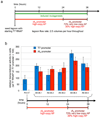
Laboratory evolution has generated many biomolecules with desired properties, but a single round of mutation, gene expression, screening or selection, and replication typically requires days or longer with frequent human intervention. Because evolutionary success is dependent on the total number of rounds performed, a means of performing laboratory evolution continuously and rapidly could dramatically enhance its effectiveness. Although researchers have accelerated individual steps in the evolutionary cycle, the only previous example of continuous directed evolution was the landmark study of Wright and Joyce, who continuously evolved RNA ligase ribozymes with an in vitro replication cycle that unfortunately cannot be easily adapted to other biomolecules. Here we describe a system that enables the continuous directed evolution of gene-encoded molecules that can be linked to protein production in Escherichia coli. During phage-assisted continuous evolution (PACE), evolving genes are transferred from host cell to host cell through a modified bacteriophage life cycle in a manner that is dependent on the activity of interest. Dozens of rounds of evolution can occur in a single day of PACE without human intervention. Using PACE, we evolved T7 RNA polymerase (RNAP) variants that recognize a distinct promoter, initiate transcripts with ATP instead of GTP, and initiate transcripts with CTP. In one example, PACE executed 200 rounds of protein evolution over the course of 8 days. Starting from undetectable activity levels in two of these cases, enzymes with each of the three target activities emerged in less than 1 week of PACE. In all three cases, PACE-evolved polymerase activities exceeded or were comparable to that of the wild-type T7 RNAP on its wild-type promoter, representing improvements of up to several hundred-fold. By greatly accelerating laboratory evolution, PACE may provide solutions to otherwise intractable directed evolution problems and address novel questions about molecular evolution.
Figures

References
References Cited
-
- Voigt CA, Kauffman S, Wang ZG. Rational evolutionary design: the theory of in vitro protein evolution. Advances in Protein Chemistry. 2000;55:79–160. - PubMed
References for the Online-only Methods Section
-
- Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009;6:343–345. - PubMed
-
- Ichetovkin IE, Abramochkin G, Shrader TE. Substrate recognition by the leucyl/phenylalanyl-tRNA-protein transferase. Conservation within the enzyme family and localization to the trypsin-resistant domain. J Biol Chem. 1997;272:33009–33014. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

