Roles of the human hypoxia-inducible factor (HIF)-3α variants in the hypoxia response
- PMID: 21479871
- PMCID: PMC11114783
- DOI: 10.1007/s00018-011-0679-5
Roles of the human hypoxia-inducible factor (HIF)-3α variants in the hypoxia response
Abstract
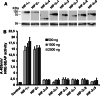
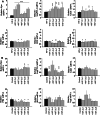
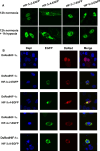
The hypoxia-inducible transcription factor (HIF) controls (in an oxygen-dependent manner) the expression of a large number of genes whose products are involved in the response of cells to hypoxia. HIF is an αβ dimer that binds to hypoxia response elements (HREs) in its target genes. Human HIF-α has three isoforms, HIF-1α, HIF-2α and HIF-3α, of which the roles of HIF-3α are largely unknown, although it is usually regarded as a negative regulator of HIF-1α and HIF-2α. The human HIF-3α locus is subject to extensive alternative splicing, leading to at least seven variants. We analyzed here the effects of the long variants and the short variant HIF-3α4 on the hypoxia response. All these variants were found to interact with HIF-β, HIF-1α and HIF-2α. The long HIF-3α variants were localized in the nucleus in hypoxia, while HIF-3α4 was cytoplasmic. Interaction of the HIF-3α variants with HIF-1α inhibited the nuclear translocation of both. None of the long HIF-3α variants was capable of efficient induction of an HRE reporter in overexpression experiments, but instead inhibited the transcriptional activation of the reporter by HIF-1 and HIF-2. Unexpectedly, siRNA knock-down of the endogenous HIF-3α variants led to downregulation of certain HIF target genes, while overexpression of individual long HIF-3α variants upregulated certain HIF target genes in a variant and target gene-specific manner under conditions in which HIF-β was not a limiting factor. These data indicate that the HIF-3α variants may have more versatile and specific roles in the regulation of the hypoxia response than previously anticipated.
Figures



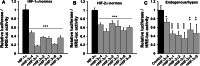

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

