Cardiac ATP-sensitive K+ channel associates with the glycolytic enzyme complex
- PMID: 21482559
- PMCID: PMC3114533
- DOI: 10.1096/fj.10-176669
Cardiac ATP-sensitive K+ channel associates with the glycolytic enzyme complex
Abstract
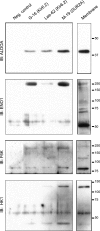
Being gated by high-energy nucleotides, cardiac ATP-sensitive potassium (K(ATP)) channels are exquisitely sensitive to changes in cellular energy metabolism. An emerging view is that proteins associated with the K(ATP) channel provide an additional layer of regulation. Using putative sulfonylurea receptor (SUR) coiled-coil domains as baits in a 2-hybrid screen against a rat cardiac cDNA library, we identified glycolytic enzymes (GAPDH and aldolase A) as putative interacting proteins. Interaction between aldolase and SUR was confirmed using GST pulldown assays and coimmunoprecipitation assays. Mass spectrometry of proteins from K(ATP) channel immunoprecipitates of rat cardiac membranes identified glycolysis as the most enriched biological process. Coimmunoprecipitation assays confirmed interaction for several glycolytic enzymes throughout the glycolytic pathway. Immunocytochemistry colocalized many of these enzymes with K(ATP) channel subunits in rat cardiac myocytes. The catalytic activities of aldolase and pyruvate kinase functionally modulate K(ATP) channels in patch-clamp experiments, whereas D-glucose was without effect. Overall, our data demonstrate close physical association and functional interaction of the glycolytic process (particularly the distal ATP-generating steps) with cardiac K(ATP) channels.
Figures



References
-
- Ashcroft F. M. (1988) Adenosine 5′-triphosphate-sensitive potassium channels. Annu. Rev. Neurosci. 11, 97–118 - PubMed
-
- Nichols C. G. (2006) KATP channels as molecular sensors of cellular metabolism. Nature 440, 470–476 - PubMed
-
- Dubinsky W. P., Mayorga-Wark O., Schultz S. G. (1998) Colocalization of glycolytic enzyme activity and KATP channels in basolateral membrane of Necturus enterocytes. Am. J. Physiol. 275, C1653–C1659 - PubMed
-
- Dhar Chowdhury P., Harrell M. D., Han S., Jankowska D., Parachuru L., Morrissey A., Srivastava S., Liu W., Malester B., Yoshida H., Coetzee W. A. (2005) The glycolytic enzymes, glyceraldehyde-3-phosphate dehydrogenase, triose phosphate isomerase and pyruvate kinase are components of the KATP channel macromolecular complex and regulate its function. J. Biol. Chem. 280, 38464–38470 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

