ECHO: a reference-free short-read error correction algorithm
- PMID: 21482625
- PMCID: PMC3129260
- DOI: 10.1101/gr.111351.110
ECHO: a reference-free short-read error correction algorithm
Abstract
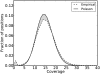
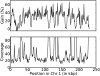
Developing accurate, scalable algorithms to improve data quality is an important computational challenge associated with recent advances in high-throughput sequencing technology. In this study, a novel error-correction algorithm, called ECHO, is introduced for correcting base-call errors in short-reads, without the need of a reference genome. Unlike most previous methods, ECHO does not require the user to specify parameters of which optimal values are typically unknown a priori. ECHO automatically sets the parameters in the assumed model and estimates error characteristics specific to each sequencing run, while maintaining a running time that is within the range of practical use. ECHO is based on a probabilistic model and is able to assign a quality score to each corrected base. Furthermore, it explicitly models heterozygosity in diploid genomes and provides a reference-free method for detecting bases that originated from heterozygous sites. On both real and simulated data, ECHO is able to improve the accuracy of previous error-correction methods by several folds to an order of magnitude, depending on the sequence coverage depth and the position in the read. The improvement is most pronounced toward the end of the read, where previous methods become noticeably less effective. Using a whole-genome yeast data set, it is demonstrated here that ECHO is capable of coping with nonuniform coverage. Also, it is shown that using ECHO to perform error correction as a preprocessing step considerably facilitates de novo assembly, particularly in the case of low-to-moderate sequence coverage depth.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
