IkappaB kinase beta promotes cell survival by antagonizing p53 functions through DeltaNp73alpha phosphorylation and stabilization
- PMID: 21482671
- PMCID: PMC3133237
- DOI: 10.1128/MCB.00964-10
IkappaB kinase beta promotes cell survival by antagonizing p53 functions through DeltaNp73alpha phosphorylation and stabilization
Abstract
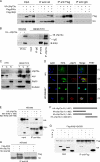
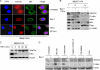
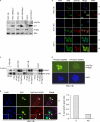
ΔNp73α, a dominant-negative inhibitor of p53 and p73, exhibits antiapoptotic and transforming activity in in vitro models and is often found to be upregulated in human cancers. The mechanisms involved in the regulation of ΔNp73α protein levels in normal and cancer cells are poorly characterized. Here, we show that that IκB kinase beta (IKKβ) increases ΔNp73α protein stability independently of its ability to activate NF-κB. IKKβ associates with and phosphorylates ΔNp73α at serine 422 (S422), leading to its accumulation in the nucleus, where it binds and represses several p53-regulated genes. S422A mutation in ΔNp73α abolished IKKβ-mediated stabilization and inhibition of p53-regulated gene expression. Inhibition of IKKβ activity by chemical inhibitors, overexpression of dominant-negative mutants, or gene silencing by siRNA also resulted in ΔNp73α destabilization, which under these conditions was rapidly translocated into the cytoplasm and degraded by a calpain-mediated mechanism. We also present evidence for the IKKβ and ΔNp73α cross talk in cancer-derived cell lines and primary cancers. Our data unveil a new mechanism involved in the regulation of the p73 and p53 network.
Figures
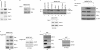






References
-
- Agami R., Blandino G., Oren M., Shaul Y. 1999. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature 399:809–813 - PubMed
-
- Anest V., et al. 2003. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature 423:659–663 - PubMed
-
- Buhlmann S., Putzer B. M. 2008. DNp73 a matter of cancer: mechanisms and clinical implications. Biochim. Biophys. Acta 1785:207–216 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
