Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones
- PMID: 21482798
- PMCID: PMC3084084
- DOI: 10.1073/pnas.1014448108
Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones
Abstract
The 70-kDa heat shock protein (Hsp70) chaperones perform a wide array of cellular functions that all derive from the ability of their N-terminal nucleotide-binding domains (NBDs) to allosterically regulate the substrate affinity of their C-terminal substrate-binding domains in a nucleotide-dependent mechanism. To explore the structural origins of Hsp70 allostery, we performed NMR analysis on the NBD of DnaK, the Escherichia coli Hsp70, in six different states (ligand-bound or apo) and in two constructs, one that retains the conserved and functionally crucial portion of the interdomain linker (residues ) and another that lacks the linker. Chemical-shift perturbation patterns identify residues at subdomain interfaces that constitute allosteric networks and enable the NBD to act as a nucleotide-modulated switch. Nucleotide binding results in changes in subdomain orientations and long-range perturbations along subdomain interfaces. In particular, our findings provide structural details for a key mechanism of Hsp70 allostery, by which information is conveyed from the nucleotide-binding site to the interdomain linker. In the presence of ATP, the linker binds to the edge of the IIA β-sheet, which structurally connects the linker and the nucleotide-binding site. Thus, a pathway of allosteric communication leads from the NBD nucleotide-binding site to the substrate-binding domain via the interdomain linker.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
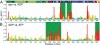
 , for backbone atoms as a function of residue number, where ΔδH, ΔδN, or ΔδCO are 1HN, 15N, and 13CO chemical-shift differences between the apo and ADP-bound states of NBD388 (A), and between the ADP- and ATP-bound states of NBD392 (B). Residues with large Δδtot (> 0.3 ppm) and significant chemical-shift perturbation (at least one ΔδH, ΔδN, or ΔδCO value is larger than two corresponding chemical-shift errors; i.e., 0.06, 0.6, and 0.6 ppm for 1HN and 15N, and 13CO atoms, respectively) are colored red and yellow, respectively; the rest are shown as cyan. The green background highlights regions that are highly affected by nucleotide binding, and the top bar shows NBD subdomains: IA (dark green), IB (light green), IIA (dark blue), IIB (light blue), crossing α-helices (red, X), the
, for backbone atoms as a function of residue number, where ΔδH, ΔδN, or ΔδCO are 1HN, 15N, and 13CO chemical-shift differences between the apo and ADP-bound states of NBD388 (A), and between the ADP- and ATP-bound states of NBD392 (B). Residues with large Δδtot (> 0.3 ppm) and significant chemical-shift perturbation (at least one ΔδH, ΔδN, or ΔδCO value is larger than two corresponding chemical-shift errors; i.e., 0.06, 0.6, and 0.6 ppm for 1HN and 15N, and 13CO atoms, respectively) are colored red and yellow, respectively; the rest are shown as cyan. The green background highlights regions that are highly affected by nucleotide binding, and the top bar shows NBD subdomains: IA (dark green), IB (light green), IIA (dark blue), IIB (light blue), crossing α-helices (red, X), the  linker motif (yellow, L), and the nucleotide-binding site (black, N).
linker motif (yellow, L), and the nucleotide-binding site (black, N).

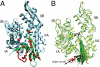
 ) bound to the hydrophobic cleft is shown as red spheres.
) bound to the hydrophobic cleft is shown as red spheres.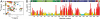
 , for backbone atoms as a function of residue number; where ΔδH, ΔδN, or ΔδCO are the largest 1HN, 15N, and 13CO chemical-shift differences between any 2 of the 12 states of NBD388 and NBD392. Coloring and the top bar are the same as for Fig. 1.
, for backbone atoms as a function of residue number; where ΔδH, ΔδN, or ΔδCO are the largest 1HN, 15N, and 13CO chemical-shift differences between any 2 of the 12 states of NBD388 and NBD392. Coloring and the top bar are the same as for Fig. 1.
Similar articles
-
An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones.Cell. 2012 Dec 7;151(6):1296-307. doi: 10.1016/j.cell.2012.11.002. Cell. 2012. PMID: 23217711 Free PMC article.
-
New insights into the structure and function of the complex between the Escherichia coli Hsp70, DnaK, and its nucleotide-exchange factor, GrpE.J Biol Chem. 2024 Jan;300(1):105574. doi: 10.1016/j.jbc.2023.105574. Epub 2023 Dec 16. J Biol Chem. 2024. PMID: 38110031 Free PMC article.
-
The Hsp70 interdomain linker is a dynamic switch that enables allosteric communication between two structured domains.J Biol Chem. 2017 Sep 8;292(36):14765-14774. doi: 10.1074/jbc.M117.789313. Epub 2017 Jul 28. J Biol Chem. 2017. PMID: 28754691 Free PMC article.
-
The Link That Binds: The Linker of Hsp70 as a Helm of the Protein's Function.Biomolecules. 2019 Sep 27;9(10):543. doi: 10.3390/biom9100543. Biomolecules. 2019. PMID: 31569820 Free PMC article. Review.
-
Intra-molecular pathways of allosteric control in Hsp70s.Philos Trans R Soc Lond B Biol Sci. 2018 Jun 19;373(1749):20170183. doi: 10.1098/rstb.2017.0183. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 29735737 Free PMC article. Review.
Cited by
-
Isoform-selective Genetic Inhibition of Constitutive Cytosolic Hsp70 Activity Promotes Client Tau Degradation Using an Altered Co-chaperone Complement.J Biol Chem. 2015 May 22;290(21):13115-27. doi: 10.1074/jbc.M115.637595. Epub 2015 Apr 11. J Biol Chem. 2015. PMID: 25864199 Free PMC article.
-
Proteasome allostery as a population shift between interchanging conformers.Proc Natl Acad Sci U S A. 2012 Dec 11;109(50):E3454-62. doi: 10.1073/pnas.1213640109. Epub 2012 Nov 12. Proc Natl Acad Sci U S A. 2012. PMID: 23150576 Free PMC article.
-
Disrupted Hydrogen-Bond Network and Impaired ATPase Activity in an Hsc70 Cysteine Mutant.Biochemistry. 2018 Feb 20;57(7):1073-1086. doi: 10.1021/acs.biochem.7b01005. Epub 2018 Feb 1. Biochemistry. 2018. PMID: 29300467 Free PMC article.
-
NMR Methods to Study Dynamic Allostery.PLoS Comput Biol. 2016 Mar 10;12(3):e1004620. doi: 10.1371/journal.pcbi.1004620. eCollection 2016 Mar. PLoS Comput Biol. 2016. PMID: 26964042 Free PMC article. Review.
-
Mechanisms of amyloid formation revealed by solution NMR.Prog Nucl Magn Reson Spectrosc. 2015 Aug;88-89:86-104. doi: 10.1016/j.pnmrs.2015.05.002. Epub 2015 May 27. Prog Nucl Magn Reson Spectrosc. 2015. PMID: 26282197 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

