Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones
- PMID: 21482798
- PMCID: PMC3084084
- DOI: 10.1073/pnas.1014448108
Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones
Abstract
The 70-kDa heat shock protein (Hsp70) chaperones perform a wide array of cellular functions that all derive from the ability of their N-terminal nucleotide-binding domains (NBDs) to allosterically regulate the substrate affinity of their C-terminal substrate-binding domains in a nucleotide-dependent mechanism. To explore the structural origins of Hsp70 allostery, we performed NMR analysis on the NBD of DnaK, the Escherichia coli Hsp70, in six different states (ligand-bound or apo) and in two constructs, one that retains the conserved and functionally crucial portion of the interdomain linker (residues ) and another that lacks the linker. Chemical-shift perturbation patterns identify residues at subdomain interfaces that constitute allosteric networks and enable the NBD to act as a nucleotide-modulated switch. Nucleotide binding results in changes in subdomain orientations and long-range perturbations along subdomain interfaces. In particular, our findings provide structural details for a key mechanism of Hsp70 allostery, by which information is conveyed from the nucleotide-binding site to the interdomain linker. In the presence of ATP, the linker binds to the edge of the IIA β-sheet, which structurally connects the linker and the nucleotide-binding site. Thus, a pathway of allosteric communication leads from the NBD nucleotide-binding site to the substrate-binding domain via the interdomain linker.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
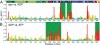
 , for backbone atoms as a function of residue number, where ΔδH, ΔδN, or ΔδCO are 1HN, 15N, and 13CO chemical-shift differences between the apo and ADP-bound states of NBD388 (A), and between the ADP- and ATP-bound states of NBD392 (B). Residues with large Δδtot (> 0.3 ppm) and significant chemical-shift perturbation (at least one ΔδH, ΔδN, or ΔδCO value is larger than two corresponding chemical-shift errors; i.e., 0.06, 0.6, and 0.6 ppm for 1HN and 15N, and 13CO atoms, respectively) are colored red and yellow, respectively; the rest are shown as cyan. The green background highlights regions that are highly affected by nucleotide binding, and the top bar shows NBD subdomains: IA (dark green), IB (light green), IIA (dark blue), IIB (light blue), crossing α-helices (red, X), the
, for backbone atoms as a function of residue number, where ΔδH, ΔδN, or ΔδCO are 1HN, 15N, and 13CO chemical-shift differences between the apo and ADP-bound states of NBD388 (A), and between the ADP- and ATP-bound states of NBD392 (B). Residues with large Δδtot (> 0.3 ppm) and significant chemical-shift perturbation (at least one ΔδH, ΔδN, or ΔδCO value is larger than two corresponding chemical-shift errors; i.e., 0.06, 0.6, and 0.6 ppm for 1HN and 15N, and 13CO atoms, respectively) are colored red and yellow, respectively; the rest are shown as cyan. The green background highlights regions that are highly affected by nucleotide binding, and the top bar shows NBD subdomains: IA (dark green), IB (light green), IIA (dark blue), IIB (light blue), crossing α-helices (red, X), the  linker motif (yellow, L), and the nucleotide-binding site (black, N).
linker motif (yellow, L), and the nucleotide-binding site (black, N).

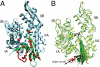
 ) bound to the hydrophobic cleft is shown as red spheres.
) bound to the hydrophobic cleft is shown as red spheres.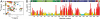
 , for backbone atoms as a function of residue number; where ΔδH, ΔδN, or ΔδCO are the largest 1HN, 15N, and 13CO chemical-shift differences between any 2 of the 12 states of NBD388 and NBD392. Coloring and the top bar are the same as for Fig. 1.
, for backbone atoms as a function of residue number; where ΔδH, ΔδN, or ΔδCO are the largest 1HN, 15N, and 13CO chemical-shift differences between any 2 of the 12 states of NBD388 and NBD392. Coloring and the top bar are the same as for Fig. 1.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

