Daxx-beta and Daxx-gamma, two novel splice variants of the transcriptional co-repressor Daxx
- PMID: 21482821
- PMCID: PMC3103337
- DOI: 10.1074/jbc.M110.196311
Daxx-beta and Daxx-gamma, two novel splice variants of the transcriptional co-repressor Daxx
Abstract
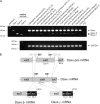
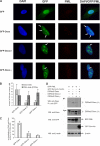
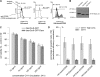
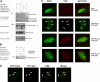
Daxx is involved in transcriptional control and apoptosis. It comprises several domains, including a regulatory C terminus that is responsible for the interaction with numerous proteins such as p53, promyelocytic leukemia protein (PML), and Hsp27. Here, we describe the identification and characterization of two novel variants of Daxx termed Daxx-β and Daxx-γ, which are generated by alternative splicing. Alternative splicing results in a truncated regulatory C terminus in both proteins. As a consequence, Daxx-β and Daxx-γ show a markedly decreased affinity to PML, which in turn is associated with a different subnuclear localization of these proteins compared with Daxx. Although Daxx is localized mainly in PML-oncogenic domains (PODs) Daxx-β and Daxx-γ display a distinct distribution pattern. Furthermore, Daxx-β and Daxx-γ show a decreased affinity to p53 also due to the truncated C terminus. We provide evidence that the p53 recruitment into PODs is Daxx isoform-dependent. The decreased affinity of Daxx-β/-γ to p53 and PML results in a diffuse localization of p53 throughout the nucleus. In contrast to Daxx, Daxx-β and Daxx-γ are unable to repress p53-mediated transcription. Therefore, alternative splicing of Daxx might indicate an additional level in the cellular apoptosis network.
Figures




Similar articles
-
Identification of Daxx interacting with p73, one of the p53 family, and its regulation of p53 activity by competitive interaction with PML.Nucleic Acids Res. 2003 Sep 15;31(18):5356-67. doi: 10.1093/nar/gkg741. Nucleic Acids Res. 2003. PMID: 12954772 Free PMC article.
-
Promyelocytic leukemia protein (PML) functions as a glucocorticoid receptor co-activator by sequestering Daxx to the PML oncogenic domains (PODs) to enhance its transactivation potential.J Biol Chem. 2003 May 2;278(18):15958-65. doi: 10.1074/jbc.M300387200. Epub 2003 Feb 20. J Biol Chem. 2003. PMID: 12595526
-
AXIN is an essential co-activator for the promyelocytic leukemia protein in p53 activation.Oncogene. 2011 Mar 10;30(10):1194-204. doi: 10.1038/onc.2010.499. Epub 2010 Nov 8. Oncogene. 2011. PMID: 21057547
-
Role of nuclear bodies in apoptosis signalling.Biochim Biophys Acta. 2008 Nov;1783(11):2185-94. doi: 10.1016/j.bbamcr.2008.07.002. Epub 2008 Jul 16. Biochim Biophys Acta. 2008. PMID: 18680765 Review.
-
PML NBs (ND10) and Daxx: from nuclear structure to protein function.Front Biosci. 2008 May 1;13:7132-42. doi: 10.2741/3216. Front Biosci. 2008. PMID: 18508722 Review.
Cited by
-
Tale of a tegument transactivator: the past, present and future of human CMV pp71.Future Virol. 2012 Sep 1;7(9):855-869. doi: 10.2217/fvl.12.86. Future Virol. 2012. PMID: 23378857 Free PMC article.
-
Tumor suppressor protein Pdcd4 interacts with Daxx and modulates the stability of Daxx and the Hipk2-dependent phosphorylation of p53 at serine 46.Oncogenesis. 2013 Jan 14;2(1):e37. doi: 10.1038/oncsis.2012.37. Oncogenesis. 2013. PMID: 23536002 Free PMC article.
-
Ubiquitin-independent proteasomal degradation of tumor suppressors by human cytomegalovirus pp71 requires the 19S regulatory particle.J Virol. 2013 Apr;87(8):4665-71. doi: 10.1128/JVI.03301-12. Epub 2013 Feb 13. J Virol. 2013. PMID: 23408605 Free PMC article.
-
Nuclear domain 10 of the viral aspect.World J Virol. 2013 Aug 12;2(3):110-22. doi: 10.5501/wjv.v2.i3.110. World J Virol. 2013. PMID: 24255882 Free PMC article. Review.
-
Death domain-associated protein (DAXX) expression is associated with poor survival in metastatic high-grade serous carcinoma.Virchows Arch. 2020 Dec;477(6):857-864. doi: 10.1007/s00428-020-02842-4. Epub 2020 Jun 12. Virchows Arch. 2020. PMID: 32533344 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

