Reconciliation of genome-scale metabolic reconstructions for comparative systems analysis
- PMID: 21483480
- PMCID: PMC3068926
- DOI: 10.1371/journal.pcbi.1001116
Reconciliation of genome-scale metabolic reconstructions for comparative systems analysis
Abstract
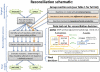
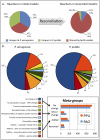
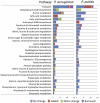
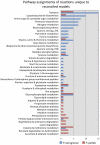
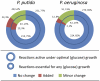
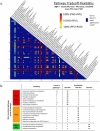
In the past decade, over 50 genome-scale metabolic reconstructions have been built for a variety of single- and multi- cellular organisms. These reconstructions have enabled a host of computational methods to be leveraged for systems-analysis of metabolism, leading to greater understanding of observed phenotypes. These methods have been sparsely applied to comparisons between multiple organisms, however, due mainly to the existence of differences between reconstructions that are inherited from the respective reconstruction processes of the organisms to be compared. To circumvent this obstacle, we developed a novel process, termed metabolic network reconciliation, whereby non-biological differences are removed from genome-scale reconstructions while keeping the reconstructions as true as possible to the underlying biological data on which they are based. This process was applied to two organisms of great importance to disease and biotechnological applications, Pseudomonas aeruginosa and Pseudomonas putida, respectively. The result is a pair of revised genome-scale reconstructions for these organisms that can be analyzed at a systems level with confidence that differences are indicative of true biological differences (to the degree that is currently known), rather than artifacts of the reconstruction process. The reconstructions were re-validated with various experimental data after reconciliation. With the reconciled and validated reconstructions, we performed a genome-wide comparison of metabolic flexibility between P. aeruginosa and P. putida that generated significant new insight into the underlying biology of these important organisms. Through this work, we provide a novel methodology for reconciling models, present new genome-scale reconstructions of P. aeruginosa and P. putida that can be directly compared at a network level, and perform a network-wide comparison of the two species. These reconstructions provide fresh insights into the metabolic similarities and differences between these important Pseudomonads, and pave the way towards full comparative analysis of genome-scale metabolic reconstructions of multiple species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Murray TS, Egan M, Kazmierczak BI. Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr Opin Pediatr. 2007;19:83–88. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

