Core gene set as the basis of multilocus sequence analysis of the subclass Actinobacteridae
- PMID: 21483493
- PMCID: PMC3069002
- DOI: 10.1371/journal.pone.0014792
Core gene set as the basis of multilocus sequence analysis of the subclass Actinobacteridae
Abstract
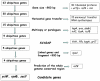
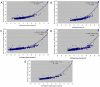
Comparative genomic sequencing is shedding new light on bacterial identification, taxonomy and phylogeny. An in silico assessment of a core gene set necessary for cellular functioning was made to determine a consensus set of genes that would be useful for the identification, taxonomy and phylogeny of the species belonging to the subclass Actinobacteridae which contained two orders Actinomycetales and Bifidobacteriales. The subclass Actinobacteridae comprised about 85% of the actinobacteria families. The following recommended criteria were used to establish a comprehensive gene set; the gene should (i) be long enough to contain phylogenetically useful information, (ii) not be subject to horizontal gene transfer, (iii) be a single copy (iv) have at least two regions sufficiently conserved that allow the design of amplification and sequencing primers and (v) predict whole-genome relationships. We applied these constraints to 50 different Actinobacteridae genomes and made 1,224 pairwise comparisons of the genome conserved regions and gene fragments obtained by using Sequence VARiability Analysis Program (SVARAP), which allow designing the primers. Following a comparative statistical modeling phase, 3 gene fragments were selected, ychF, rpoB, and secY with R2>0.85. Selected sets of broad range primers were tested from the 3 gene fragments and were demonstrated to be useful for amplification and sequencing of 25 species belonging to 9 genera of Actinobacteridae. The intraspecies similarities were 96.3-100% for ychF, 97.8-100% for rpoB and 96.9-100% for secY among 73 strains belonging to 15 species of the subclass Actinobacteridae compare to 99.4-100% for 16S rRNA. The phylogenetic topology obtained from the combined datasets ychF+rpoB+secY was globally similar to that inferred from the 16S rRNA but with higher confidence. It was concluded that multi-locus sequence analysis using core gene set might represent the first consensus and valid approach for investigating the bacterial identification, phylogeny and taxonomy.
Conflict of interest statement
Figures

References
-
- Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kämpfer P, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. - PubMed
-
- Wertz JE, Goldstone C, Gordon DM Riley MA. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J Evol Biol. 2003;16:1236–1248. - PubMed
-
- Zeigler DR. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int J Syst Evol Microbiol. 2003;53:1893–1900. - PubMed
-
- Adekambi T, Drancourt M. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int J Syst Evol Microbiol. 2004;54:2095–2105. - PubMed
-
- Devulder G, Pérouse de Montclos M, Flandrois JP. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int J Syst Evol Microbiol. 2005;55:293–302. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

