β-catenin accumulation in nuclei of hepatocellular carcinoma cells up-regulates glutathione-s-transferase M3 mRNA
- PMID: 21483640
- PMCID: PMC3072644
- DOI: 10.3748/wjg.v17.i13.1772
β-catenin accumulation in nuclei of hepatocellular carcinoma cells up-regulates glutathione-s-transferase M3 mRNA
Abstract
Aim: To identify the differentially over-expressed genes associated with β-catenin accumulation in nuclei of hepatocellular carcinoma (HCC) cells.
Methods: Differentially expressed genes were identified in radiation-induced B6C3 F1 mouse HCC cells by mRNA differential display, Northern blot and RT-PCR, respectively. Total glutathione-s-transferase (GST) activity was measured by GST activity assay and β-catenin localization was detected with immunostaining in radiation-induced mouse HCC cells and in HepG2 cell lines.
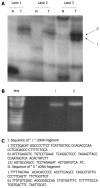
Results: Two up-regulated genes, glutamine synthetase and glutathione-s-transferase M3 (GSTM3), were identified in radiation-induced mouse HCC cells. Influence of β-catenin accumulation in nuclei of HCC cells on up-regulation of GSTM3 mRNA was investigated. The nearby upstream domain of GSTM3 contained the β-catenin/Tcf-Lef consensus binding site sequences [5'-(A/T)(A/T) CAAAG-3'], and the total GST activity ratio was considerably higher in B6C3F1 mouse HCC cells with β-catenin accumulation in nuclei of HCC cells than in those without β-catenin accumulation (0.353 ± 0.117 vs. 0.071 ± 0.064, P < 0.001). The TWS119 (a distinct GSK-3β inhibitor)-induced total GST activity was significantly higher in HepG2 cells with β-catenin accumulation than in those without β-catenin accumulation in nuclei of HCC cells. Additionally, the GSTM3 mRNA level was significantly higher at 24 h than at 12 h in TWS119-treated HepG2 cells.
Conclusion: β-catenin accumulation increases GST activity in nuclei of HCC cells, and GSTM3 may be a novel target gene of the β-catenin/Tcf-Lef complex.
Keywords: Differential display analysis; Glutathione-s-transferase M3; Hepatocellular carcinoma; Radiation; β-catenin accumulation.
Figures

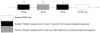
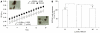
References
-
- Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. - PubMed
-
- Takada S, Koike K. Activated N-ras gene was found in human hepatoma tissue but only in a small fraction of the tumor cells. Oncogene. 1989;4:189–193. - PubMed
-
- Calvisi DF, Thorgeirsson SS. Molecular mechanisms of hepatocarcinogenesis in transgenic mouse models of liver cancer. Toxicol Pathol. 2005;33:181–184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

