Identification of common differentially expressed genes in urinary bladder cancer
- PMID: 21483740
- PMCID: PMC3070717
- DOI: 10.1371/journal.pone.0018135
Identification of common differentially expressed genes in urinary bladder cancer
Abstract
Background: Current diagnosis and treatment of urinary bladder cancer (BC) has shown great progress with the utilization of microarrays.
Purpose: Our goal was to identify common differentially expressed (DE) genes among clinically relevant subclasses of BC using microarrays.
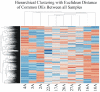
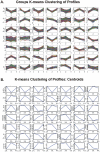
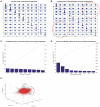
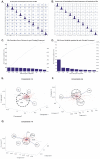
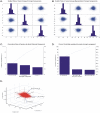
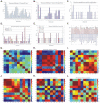
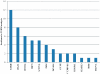
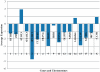
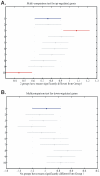
Methodology/principal findings: BC samples and controls, both experimental and publicly available datasets, were analyzed by whole genome microarrays. We grouped the samples according to their histology and defined the DE genes in each sample individually, as well as in each tumor group. A dual analysis strategy was followed. First, experimental samples were analyzed and conclusions were formulated; and second, experimental sets were combined with publicly available microarray datasets and were further analyzed in search of common DE genes. The experimental dataset identified 831 genes that were DE in all tumor samples, simultaneously. Moreover, 33 genes were up-regulated and 85 genes were down-regulated in all 10 BC samples compared to the 5 normal tissues, simultaneously. Hierarchical clustering partitioned tumor groups in accordance to their histology. K-means clustering of all genes and all samples, as well as clustering of tumor groups, presented 49 clusters. K-means clustering of common DE genes in all samples revealed 24 clusters. Genes manifested various differential patterns of expression, based on PCA. YY1 and NFκB were among the most common transcription factors that regulated the expression of the identified DE genes. Chromosome 1 contained 32 DE genes, followed by chromosomes 2 and 11, which contained 25 and 23 DE genes, respectively. Chromosome 21 had the least number of DE genes. GO analysis revealed the prevalence of transport and binding genes in the common down-regulated DE genes; the prevalence of RNA metabolism and processing genes in the up-regulated DE genes; as well as the prevalence of genes responsible for cell communication and signal transduction in the DE genes that were down-regulated in T1-Grade III tumors and up-regulated in T2/T3-Grade III tumors. Combination of samples from all microarray platforms revealed 17 common DE genes, (BMP4, CRYGD, DBH, GJB1, KRT83, MPZ, NHLH1, TACR3, ACTC1, MFAP4, SPARCL1, TAGLN, TPM2, CDC20, LHCGR, TM9SF1 and HCCS) 4 of which participate in numerous pathways.
Conclusions/significance: The identification of the common DE genes among BC samples of different histology can provide further insight into the discovery of new putative markers.
Conflict of interest statement
Figures










References
-
- Lopez-Beltran A, Montironi R. Non-invasive urothelial neoplasms: according to the most recent WHO classification. Eur Urol. 2004;46:170–176. - PubMed
-
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. - PubMed
-
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

