Eukaryotic richness in the abyss: insights from pyrotag sequencing
- PMID: 21483744
- PMCID: PMC3070721
- DOI: 10.1371/journal.pone.0018169
Eukaryotic richness in the abyss: insights from pyrotag sequencing
Abstract
Background: The deep sea floor is considered one of the most diverse ecosystems on Earth. Recent environmental DNA surveys based on clone libraries of rRNA genes confirm this observation and reveal a high diversity of eukaryotes present in deep-sea sediment samples. However, environmental clone-library surveys yield only a modest number of sequences with which to evaluate the diversity of abyssal eukaryotes.
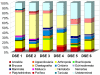
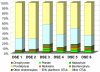
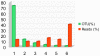
Methodology/principal findings: Here, we examined the richness of eukaryotic DNA in deep Arctic and Southern Ocean samples using massively parallel sequencing of the 18S ribosomal RNA (rRNA) V9 hypervariable region. In very small volumes of sediments, ranging from 0.35 to 0.7 g, we recovered up to 7,499 unique sequences per sample. By clustering sequences having up to 3 differences, we observed from 942 to 1756 Operational Taxonomic Units (OTUs) per sample. Taxonomic analyses of these OTUs showed that DNA of all major groups of eukaryotes is represented at the deep-sea floor. The dinoflagellates, cercozoans, ciliates, and euglenozoans predominate, contributing to 17%, 16%, 10%, and 8% of all assigned OTUs, respectively. Interestingly, many sequences represent photosynthetic taxa or are similar to those reported from the environmental surveys of surface waters. Moreover, each sample contained from 31 to 71 different metazoan OTUs despite the small sample volume collected. This indicates that a significant faction of the eukaryotic DNA sequences likely do not belong to living organisms, but represent either free, extracellular DNA or remains and resting stages of planktonic species.
Conclusions/significance: In view of our study, the deep-sea floor appears as a global DNA repository, which preserves genetic information about organisms living in the sediment, as well as in the water column above it. This information can be used for future monitoring of past and present environmental changes.
Conflict of interest statement
Figures


Similar articles
-
Censusing marine eukaryotic diversity in the twenty-first century.Philos Trans R Soc Lond B Biol Sci. 2016 Sep 5;371(1702):20150331. doi: 10.1098/rstb.2015.0331. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27481783 Free PMC article. Review.
-
Investigating microbial eukaryotic diversity from a global census: insights from a comparison of pyrotag and full-length sequences of 18S rRNA genes.Appl Environ Microbiol. 2014 Jul;80(14):4363-73. doi: 10.1128/AEM.00057-14. Epub 2014 May 9. Appl Environ Microbiol. 2014. PMID: 24814788 Free PMC article.
-
Ciliate diversity and distribution patterns in the sediments of a seamount and adjacent abyssal plains in the tropical Western Pacific Ocean.BMC Microbiol. 2017 Sep 12;17(1):192. doi: 10.1186/s12866-017-1103-6. BMC Microbiol. 2017. PMID: 28899339 Free PMC article.
-
Ultra-deep sequencing of foraminiferal microbarcodes unveils hidden richness of early monothalamous lineages in deep-sea sediments.Proc Natl Acad Sci U S A. 2011 Aug 9;108(32):13177-82. doi: 10.1073/pnas.1018426108. Epub 2011 Jul 25. Proc Natl Acad Sci U S A. 2011. PMID: 21788523 Free PMC article.
-
Unveiling new microbial eukaryotes in the surface ocean.Curr Opin Microbiol. 2008 Jun;11(3):213-8. doi: 10.1016/j.mib.2008.04.004. Epub 2008 Jun 13. Curr Opin Microbiol. 2008. PMID: 18556239 Review.
Cited by
-
High and specific diversity of protists in the deep-sea basins dominated by diplonemids, kinetoplastids, ciliates and foraminiferans.Commun Biol. 2021 Apr 23;4(1):501. doi: 10.1038/s42003-021-02012-5. Commun Biol. 2021. PMID: 33893386 Free PMC article.
-
Patterns and Drivers of Vertical Distribution of the Ciliate Community from the Surface to the Abyssopelagic Zone in the Western Pacific Ocean.Front Microbiol. 2017 Dec 19;8:2559. doi: 10.3389/fmicb.2017.02559. eCollection 2017. Front Microbiol. 2017. PMID: 29312240 Free PMC article.
-
Molecular diversity of bacteria in commercially available "Spirulina" food supplements.PeerJ. 2016 Jan 21;4:e1610. doi: 10.7717/peerj.1610. eCollection 2016. PeerJ. 2016. PMID: 26819852 Free PMC article.
-
Censusing marine eukaryotic diversity in the twenty-first century.Philos Trans R Soc Lond B Biol Sci. 2016 Sep 5;371(1702):20150331. doi: 10.1098/rstb.2015.0331. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27481783 Free PMC article. Review.
-
Improved 18S and 28S rDNA primer sets for NGS-based parasite detection.Sci Rep. 2019 Oct 31;9(1):15789. doi: 10.1038/s41598-019-52422-z. Sci Rep. 2019. PMID: 31673037 Free PMC article.
References
-
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. - PubMed
-
- Creer S, Fonseca VG, Porazinska DL, Giblin-Davis RM, Sung W, et al. Ultrasequencing of the meiofaunal biosphere: practice, pitfalls and promises. Mol Ecol. 2010;19:4–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

