Genome-wide expression profiling of five mouse models identifies similarities and differences with human psoriasis
- PMID: 21483750
- PMCID: PMC3070727
- DOI: 10.1371/journal.pone.0018266
Genome-wide expression profiling of five mouse models identifies similarities and differences with human psoriasis
Abstract
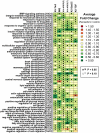
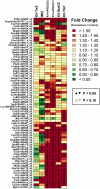
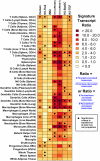
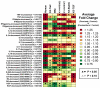
Development of a suitable mouse model would facilitate the investigation of pathomechanisms underlying human psoriasis and would also assist in development of therapeutic treatments. However, while many psoriasis mouse models have been proposed, no single model recapitulates all features of the human disease, and standardized validation criteria for psoriasis mouse models have not been widely applied. In this study, whole-genome transcriptional profiling is used to compare gene expression patterns manifested by human psoriatic skin lesions with those that occur in five psoriasis mouse models (K5-Tie2, imiquimod, K14-AREG, K5-Stat3C and K5-TGFbeta1). While the cutaneous gene expression profiles associated with each mouse phenotype exhibited statistically significant similarity to the expression profile of psoriasis in humans, each model displayed distinctive sets of similarities and differences in comparison to human psoriasis. For all five models, correspondence to the human disease was strong with respect to genes involved in epidermal development and keratinization. Immune and inflammation-associated gene expression, in contrast, was more variable between models as compared to the human disease. These findings support the value of all five models as research tools, each with identifiable areas of convergence to and divergence from the human disease. Additionally, the approach used in this paper provides an objective and quantitative method for evaluation of proposed mouse models of psoriasis, which can be strategically applied in future studies to score strengths of mouse phenotypes relative to specific aspects of human psoriasis.
Conflict of interest statement
Figures

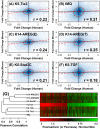

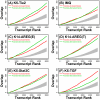
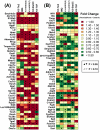
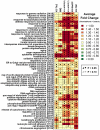
References
-
- Conrad C, Boyman O, Tonel G, Tun-Kyi A, Laggner U, et al. Alpha1beta1 integrin is crucial for accumulation of epidermal T cells and the development of psoriasis. Nat Med. 2007;13:836–842. - PubMed
-
- Boehncke WH, Schön MP. Animal models of psoriasis. Clin Dermatol. 2007;25:596–605. - PubMed
-
- Gudjonsson JE, Johnston A, Dyson M, Valdimarsson H, Elder JT. Mouse models of psoriasis. J Invest Dermatol. 2007;127:1292–1308. - PubMed
-
- Nestle FO, Nickoloff BJ. Animal models of psoriasis: a brief update. J Eur Acad Dermatol Venereol. 2006;20:24–27.
Publication types
MeSH terms
Substances
Grants and funding
- CA79998/CA/NCI NIH HHS/United States
- P30 AR039750/AR/NIAMS NIH HHS/United States
- CA76520/CA/NCI NIH HHS/United States
- R01 GM070966/GM/NIGMS NIH HHS/United States
- P50 AR055508/AR/NIAMS NIH HHS/United States
- AR054966/AR/NIAMS NIH HHS/United States
- R01 AR054966/AR/NIAMS NIH HHS/United States
- AR052889/AR/NIAMS NIH HHS/United States
- R01 CA079998/CA/NCI NIH HHS/United States
- GM70966/GM/NIGMS NIH HHS/United States
- CA89849/CA/NCI NIH HHS/United States
- P30AR39750/AR/NIAMS NIH HHS/United States
- R01 CA076520/CA/NCI NIH HHS/United States
- R01 AR052889/AR/NIAMS NIH HHS/United States
- P50AR05508/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

