REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock
- PMID: 21483796
- PMCID: PMC3069099
- DOI: 10.1371/journal.pgen.1001350
REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock
Abstract
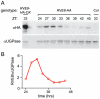
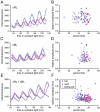
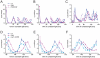
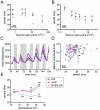
Circadian rhythms provide organisms with an adaptive advantage, allowing them to regulate physiological and developmental events so that they occur at the most appropriate time of day. In plants, as in other eukaryotes, multiple transcriptional feedback loops are central to clock function. In one such feedback loop, the Myb-like transcription factors CCA1 and LHY directly repress expression of the pseudoresponse regulator TOC1 by binding to an evening element (EE) in the TOC1 promoter. Another key regulatory circuit involves CCA1 and LHY and the TOC1 homologs PRR5, PRR7, and PRR9. Purification of EE-binding proteins from plant extracts followed by mass spectrometry led to the identification of RVE8, a homolog of CCA1 and LHY. Similar to these well-known clock genes, expression of RVE8 is circadian-regulated with a dawn phase of expression, and RVE8 binds specifically to the EE. However, whereas cca1 and lhy mutants have short period phenotypes and overexpression of either gene causes arrhythmia, rve8 mutants have long-period and RVE8-OX plants have short-period phenotypes. Light input to the clock is normal in rve8, but temperature compensation (a hallmark of circadian rhythms) is perturbed. RVE8 binds to the promoters of both TOC1 and PRR5 in the subjective afternoon, but surprisingly only PRR5 expression is perturbed by overexpression of RVE8. Together, our data indicate that RVE8 promotes expression of a subset of EE-containing clock genes towards the end of the subjective day and forms a negative feedback loop with PRR5. Thus RVE8 and its homologs CCA1 and LHY function close to the circadian oscillator but act via distinct molecular mechanisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

