H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation
- PMID: 21483810
- PMCID: PMC3069113
- DOI: 10.1371/journal.pgen.1001354
H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation
Abstract
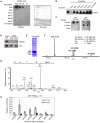
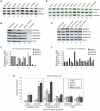
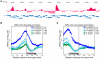
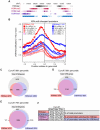
Methylation of histone H3 lysine 4 (H3K4me) is an evolutionarily conserved modification whose role in the regulation of gene expression has been extensively studied. In contrast, the function of H3K4 acetylation (H3K4ac) has received little attention because of a lack of tools to separate its function from that of H3K4me. Here we show that, in addition to being methylated, H3K4 is also acetylated in budding yeast. Genetic studies reveal that the histone acetyltransferases (HATs) Gcn5 and Rtt109 contribute to H3K4 acetylation in vivo. Whilst removal of H3K4ac from euchromatin mainly requires the histone deacetylase (HDAC) Hst1, Sir2 is needed for H3K4 deacetylation in heterochomatin. Using genome-wide chromatin immunoprecipitation (ChIP), we show that H3K4ac is enriched at promoters of actively transcribed genes and located just upstream of H3K4 tri-methylation (H3K4me3), a pattern that has been conserved in human cells. We find that the Set1-containing complex (COMPASS), which promotes H3K4me2 and -me3, also serves to limit the abundance of H3K4ac at gene promoters. In addition, we identify a group of genes that have high levels of H3K4ac in their promoters and are inadequately expressed in H3-K4R, but not in set1Δ mutant strains, suggesting that H3K4ac plays a positive role in transcription. Our results reveal a novel regulatory feature of promoter-proximal chromatin, involving mutually exclusive histone modifications of the same histone residue (H3K4ac and H3K4me).
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. - PubMed
-
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, et al. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. - PubMed
-
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

