DNA methylation, the early-life social environment and behavioral disorders
- PMID: 21484196
- PMCID: PMC3261271
- DOI: 10.1007/s11689-011-9079-2
DNA methylation, the early-life social environment and behavioral disorders
Abstract
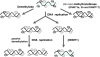
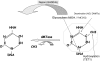
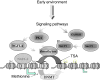
One of the outstanding questions in behavioral disorders is untangling the complex relationship between nurture and nature. Although epidemiological data provide evidence that there is an interaction between genetics (nature) and the social and physical environments (nurture) in a spectrum of behavioral disorders, the main open question remains the mechanism. Emerging data support the hypothesis that DNA methylation, a covalent modification of the DNA molecule that is a component of its chemical structure, serves as an interface between the dynamic environment and the fixed genome. We propose that modulation of DNA methylation in response to environmental cues early in life serves as a mechanism of life-long genome adaptation. Under certain contexts, this adaptation can turn maladaptive resulting in behavioral disorders. This hypothesis has important implications on understanding, predicting, preventing, and treating behavioral disorders including autism that will be discussed.
Figures
Similar articles
-
The early life social environment and DNA methylation: DNA methylation mediating the long-term impact of social environments early in life.Epigenetics. 2011 Aug;6(8):971-8. doi: 10.4161/epi.6.8.16793. Epub 2011 Aug 1. Epigenetics. 2011. PMID: 21772123 Free PMC article. Review.
-
The early-life social environment and DNA methylation.Clin Genet. 2012 Apr;81(4):341-9. doi: 10.1111/j.1399-0004.2012.01843.x. Epub 2012 Feb 13. Clin Genet. 2012. PMID: 22236068 Review.
-
Mind-body interrelationship in DNA methylation.Chem Immunol Allergy. 2012;98:85-99. doi: 10.1159/000336503. Epub 2012 Jun 26. Chem Immunol Allergy. 2012. PMID: 22767059
-
The genome- and system-wide response of DNA methylation to early life adversity and its implication on mental health.Can J Psychiatry. 2013 Dec;58(12):697-704. doi: 10.1177/070674371305801208. Can J Psychiatry. 2013. PMID: 24331290 Review.
-
DNA methylation, behavior and early life adversity.J Genet Genomics. 2013 Jul 20;40(7):331-8. doi: 10.1016/j.jgg.2013.06.004. Epub 2013 Jun 25. J Genet Genomics. 2013. PMID: 23876773 Review.
Cited by
-
Tell me what you eat and I will tell you your sociotype: coping with diabesity.Rambam Maimonides Med J. 2012 Apr 30;3(2):e0010. doi: 10.5041/RMMJ.10077. Print 2012 Apr. Rambam Maimonides Med J. 2012. PMID: 23908834 Free PMC article.
-
DRDs and Brain-Derived Neurotrophic Factor Share a Common Therapeutic Ground: A Novel Bioinformatic Approach Sheds New Light Toward Pharmacological Treatment of Cognitive and Behavioral Disorders.Adv Exp Med Biol. 2023;1424:97-115. doi: 10.1007/978-3-031-31982-2_11. Adv Exp Med Biol. 2023. PMID: 37486484
-
Pharmacological activation of group-II metabotropic glutamate receptors corrects a schizophrenia-like phenotype induced by prenatal stress in mice.Neuropsychopharmacology. 2012 Mar;37(4):929-38. doi: 10.1038/npp.2011.274. Epub 2011 Nov 16. Neuropsychopharmacology. 2012. PMID: 22089319 Free PMC article.
-
Genome-wide profiling of DNA methylome and transcriptome in peripheral blood monocytes for major depression: A Monozygotic Discordant Twin Study.Transl Psychiatry. 2019 Sep 2;9(1):215. doi: 10.1038/s41398-019-0550-2. Transl Psychiatry. 2019. PMID: 31477685 Free PMC article.
-
An epigenome-wide association study meta-analysis of educational attainment.Mol Psychiatry. 2017 Dec;22(12):1680-1690. doi: 10.1038/mp.2017.210. Epub 2017 Oct 31. Mol Psychiatry. 2017. PMID: 29086770 Free PMC article.
References
LinkOut - more resources
Full Text Sources
