An emerging model for BAP1's role in regulating cell cycle progression
- PMID: 21484256
- PMCID: PMC3128820
- DOI: 10.1007/s12013-011-9184-6
An emerging model for BAP1's role in regulating cell cycle progression
Abstract
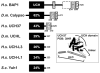
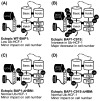
BRCA1-associated protein-1 (BAP1) is a 729 residue, nuclear-localized deubiquitinating enzyme (DUB) that displays tumor suppressor properties in the BAP1-null NCI-H226 lung carcinoma cell line. Studies that have altered BAP1 cellular levels or enzymatic activity have reported defects in cell cycle progression, notably at the G1/S transition. Recently BAP1 was shown to associate with the transcriptional regulator host cell factor 1 (HCF-1). The BAP1/HCF-1 interaction is mediated by the HCF-1 Kelch domain and an HCF-1 binding motif (HBM) within BAP1. HCF-1 is modified with ubiquitin in vivo, and ectopic studies suggest BAP1 deubiquitinates HCF-1. HCF-1 is a chromatin-associated protein thought to both activate and repress transcription by linking appropriate histone-modifying enzymes to a subset of transcription factors. One known role of HCF-1 is to promote cell cycle progression at the G1/S boundary by recruiting H3K4 histone methyltransferases to the E2F1 transcription factor so that genes required for S-phase can be transcribed. Given the robust associations between BAP1/HCF-1 and HCF-1/E2Fs, it is reasonable to speculate that BAP1 influences cell proliferation at G1/S by co-regulating transcription from HCF-1/E2F-governed promoters.
Figures
References
-
- Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. - PubMed
-
- Nishikawa H, Wu W, Koike A, Kojima R, Gomi H, Fukuda M, et al. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Research. 2009;69:111–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

