Mechlorethamine-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells
- PMID: 21486066
- PMCID: PMC3208907
- DOI: 10.1021/pr200042u
Mechlorethamine-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells
Abstract
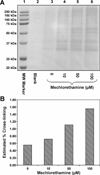
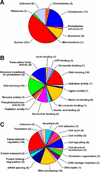
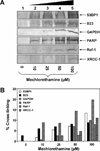
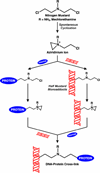
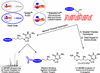
Antitumor nitrogen mustards, such as bis(2-chloroethyl)methylamine (mechlorethamine), are useful chemotherapeutic agents with a long history of clinical application. The antitumor effects of nitrogen mustards are attributed to their ability to induce DNA-DNA and DNA-protein cross-links (DPCs) that block DNA replication. In the present work, a mass spectrometry-based methodology was employed to characterize in vivo DNA-protein cross-linking following treatment of human fibrosarcoma (HT1080) cells with cytotoxic concentrations of mechlorethamine. A combination of mass spectrometry-based proteomics and immunological detection was used to identify 38 nuclear proteins that were covalently cross-linked to chromosomal DNA following treatment with mechlorethamine. Isotope dilution HPLC-ESI(+)-MS/MS analysis of total proteolytic digests revealed a concentration-dependent formation of N-[2-(S-cysteinyl)ethyl]-N-[2-(guan-7-yl)ethyl]methylamine (Cys-N7G-EMA) conjugates, indicating that mechlorethamine cross-links cysteine thiols within proteins to N-7 positions of guanine in DNA.
Figures




References
-
- Barker S, Weinfeld M, Zheng J, Li L, Murray D. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J. Biol. Chem. 2005;280:33826–33838. - PubMed
-
- Kloster M, Kostrhunova H, Zaludova R, Malina J, Kasparkova J, Brabec V, Farrell N. Trifunctional dinuclear platinum complexes as DNA-protein cross-linking agent. Biochemistry. 2004;43:7776–7786. - PubMed
-
- Ewig RA, Kohn KW. DNA-protein cross-linking and DNA interstrand cross-linking by haloethylnitrosoureas in L1210 cells. Cancer Res. 1978;38:3197–3203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

