Contribution of SecDF to Staphylococcus aureus resistance and expression of virulence factors
- PMID: 21486434
- PMCID: PMC3090319
- DOI: 10.1186/1471-2180-11-72
Contribution of SecDF to Staphylococcus aureus resistance and expression of virulence factors
Abstract
Background: SecDF is an accessory factor of the conserved Sec protein translocation machinery and belongs to the resistance-nodulation-cell division (RND) family of multidrug exporters. SecDF has been shown in Escherichia coli and Bacillus subtilis to be involved in the export of proteins. RND proteins can mediate resistance against various substances and might be of relevance in antimicrobial therapy. The role of RND proteins in Staphylococcus aureus has not yet been determined.
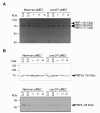
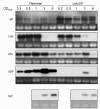
Results: Markerless deletion mutants were constructed to analyze the impact of the so far uncharacterized RND proteins in S. aureus. While the lack of Sa2056 and Sa2339 caused no phenotype regarding growth and resistance, the secDF mutant resulted in a pleiotropic phenotype. The secDF mutant was cold sensitive, but grew normally in rich medium at 37°C. Resistance to beta-lactams, glycopeptides and the RND substrates acriflavine, ethidium bromide and sodium dodecyl sulfate was reduced. The secDF mutant showed an aberrant cell separation and increased spontaneous and Triton X-100 induced autolysis, although the amounts of penicillin-binding proteins in the membrane were unchanged. The impact of secDF deletion on transcription and expression of specific virulence determinants varied: While coagulase transcription and activity were reduced, the opposite was observed for the autolysin Atl. A reduction of the transcription of the cell wall anchored protein A (spa) was also found. The accumulation of SpA in the membrane and lowered amounts in the cell wall pointed to an impaired translocation.
Conclusions: The combination of different effects of secDF deletion on transcription, regulation and translocation lead to impaired cell division, reduced resistance and altered expression of virulence determinants suggesting SecDF to be of major relevance in S. aureus. Thus SecDF could be a potential target for the control and eradication of S. aureus in the future.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

