Clostridium botulinum group III: a group with dual identity shaped by plasmids, phages and mobile elements
- PMID: 21486474
- PMCID: PMC3098183
- DOI: 10.1186/1471-2164-12-185
Clostridium botulinum group III: a group with dual identity shaped by plasmids, phages and mobile elements
Abstract
Background: Clostridium botulinum strains can be divided into four physiological groups that are sufficiently diverged to be considered as separate species. Here we present the first complete genome of a C. botulinum strain from physiological group III, causing animal botulism. We also compare the sequence to three new draft genomes from the same physiological group.
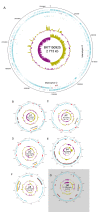
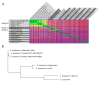
Results: The 2.77 Mb chromosome was highly conserved between the isolates and also closely related to that of C. novyi. However, the sequence was very different from the human C. botulinum group genomes. Replication-directed translocations were rare and conservation of synteny was high. The largest difference between C. botulinum group III isolates occurred within their surprisingly large plasmidomes and in the pattern of mobile elements insertions. Five plasmids, constituting 13.5% of the total genetic material, were present in the completed genome. Interestingly, the set of plasmids differed compared to other isolates. The largest plasmid, the botulinum-neurotoxin carrying prophage, was conserved at a level similar to that of the chromosome while the medium-sized plasmids seemed to be undergoing faster genetic drift. These plasmids also contained more mobile elements than other replicons. Several toxins and resistance genes were identified, many of which were located on the plasmids.
Conclusions: The completion of the genome of C. botulinum group III has revealed it to be a genome with dual identity. It belongs to the pathogenic species C. botulinum, but as a genotypic species it should also include C. novyi and C. haemolyticum. The genotypic species share a conserved chromosomal core that can be transformed into various pathogenic variants by modulation of the highly plastic plasmidome.
Figures


Similar articles
-
Plasmidome interchange between Clostridium botulinum, Clostridium novyi and Clostridium haemolyticum converts strains of independent lineages into distinctly different pathogens.PLoS One. 2014 Sep 25;9(9):e107777. doi: 10.1371/journal.pone.0107777. eCollection 2014. PLoS One. 2014. PMID: 25254374 Free PMC article.
-
Investigation of Clostridium botulinum group III's mobilome content.Anaerobe. 2018 Feb;49:71-77. doi: 10.1016/j.anaerobe.2017.12.009. Epub 2017 Dec 26. Anaerobe. 2018. PMID: 29287670
-
Pan-Genomic Analysis of Clostridium botulinum Group II (Non-Proteolytic C. botulinum) Associated with Foodborne Botulism and Isolated from the Environment.Toxins (Basel). 2020 May 8;12(5):306. doi: 10.3390/toxins12050306. Toxins (Basel). 2020. PMID: 32397147 Free PMC article.
-
Genomics of Clostridium botulinum group III strains.Res Microbiol. 2015 May;166(4):318-25. doi: 10.1016/j.resmic.2014.07.016. Epub 2014 Aug 9. Res Microbiol. 2015. PMID: 25111022 Review.
-
Biology and genomic analysis of Clostridium botulinum.Adv Microb Physiol. 2009;55:183-265, 320. doi: 10.1016/S0065-2911(09)05503-9. Adv Microb Physiol. 2009. PMID: 19573697 Review.
Cited by
-
Whole-genome single-nucleotide-polymorphism analysis for discrimination of Clostridium botulinum group I strains.Appl Environ Microbiol. 2014 Apr;80(7):2125-32. doi: 10.1128/AEM.03934-13. Epub 2014 Jan 24. Appl Environ Microbiol. 2014. PMID: 24463972 Free PMC article.
-
Evolution of Chromosomal Clostridium botulinum Type E Neurotoxin Gene Clusters: Evidence Provided by Their Rare Plasmid-Borne Counterparts.Genome Biol Evol. 2016 Mar 2;8(3):540-55. doi: 10.1093/gbe/evw017. Genome Biol Evol. 2016. PMID: 26936890 Free PMC article.
-
The Distinctive Evolution of orfX Clostridium parabotulinum Strains and Their Botulinum Neurotoxin Type A and F Gene Clusters Is Influenced by Environmental Factors and Gene Interactions via Mobile Genetic Elements.Front Microbiol. 2021 Feb 26;12:566908. doi: 10.3389/fmicb.2021.566908. eCollection 2021. Front Microbiol. 2021. PMID: 33716993 Free PMC article.
-
Navigating the Complexities Involving the Identification of Botulinum Neurotoxins (BoNTs) and the Taxonomy of BoNT-Producing Clostridia.Toxins (Basel). 2023 Sep 3;15(9):545. doi: 10.3390/toxins15090545. Toxins (Basel). 2023. PMID: 37755971 Free PMC article. Review.
-
Plasmidome interchange between Clostridium botulinum, Clostridium novyi and Clostridium haemolyticum converts strains of independent lineages into distinctly different pathogens.PLoS One. 2014 Sep 25;9(9):e107777. doi: 10.1371/journal.pone.0107777. eCollection 2014. PLoS One. 2014. PMID: 25254374 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

