Assessing the benefits of using mate-pairs to resolve repeats in de novo short-read prokaryotic assemblies
- PMID: 21486487
- PMCID: PMC3103447
- DOI: 10.1186/1471-2105-12-95
Assessing the benefits of using mate-pairs to resolve repeats in de novo short-read prokaryotic assemblies
Abstract
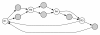
Background: Next-generation sequencing technologies allow genomes to be sequenced more quickly and less expensively than ever before. However, as sequencing technology has improved, the difficulty of de novo genome assembly has increased, due in large part to the shorter reads generated by the new technologies. The use of mated sequences (referred to as mate-pairs) is a standard means of disambiguating assemblies to obtain a more complete picture of the genome without resorting to manual finishing. Here, we examine the effectiveness of mate-pair information in resolving repeated sequences in the DNA (a paramount issue to overcome). While it has been empirically accepted that mate-pairs improve assemblies, and a variety of assemblers use mate-pairs in the context of repeat resolution, the effectiveness of mate-pairs in this context has not been systematically evaluated in previous literature.
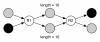
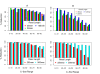
Results: We show that, in high-coverage prokaryotic assemblies, libraries of short mate-pairs (about 4-6 times the read-length) more effectively disambiguate repeat regions than the libraries that are commonly constructed in current genome projects. We also demonstrate that the best assemblies can be obtained by 'tuning' mate-pair libraries to accommodate the specific repeat structure of the genome being assembled - information that can be obtained through an initial assembly using unpaired reads. These results are shown across 360 simulations on 'ideal' prokaryotic data as well as assembly of 8 bacterial genomes using SOAPdenovo. The simulation results provide an upper-bound on the potential value of mate-pairs for resolving repeated sequences in real prokaryotic data sets. The assembly results show that our method of tuning mate-pairs exploits fundamental properties of these genomes, leading to better assemblies even when using an off -the-shelf assembler in the presence of base-call errors.
Conclusions: Our results demonstrate that dramatic improvements in prokaryotic genome assembly quality can be achieved by tuning mate-pair sizes to the actual repeat structure of a genome, suggesting the possible need to change the way sequencing projects are designed. We propose that a two-tiered approach - first generate an assembly of the genome with unpaired reads in order to evaluate the repeat structure of the genome; then generate the mate-pair libraries that provide most information towards the resolution of repeats in the genome being assembled - is not only possible, but likely also more cost-effective as it will significantly reduce downstream manual finishing costs. In future work we intend to address the question of whether this result can be extended to larger eukaryotic genomes, where repeat structure can be quite different.
Figures


References
-
- Pevzner P, Tang H. Fragment assembly with double-barreled data. Bioinformatics. 2001;17(suppl 1):S225–233. - PubMed
-
- Fleischmann R, Adams M, White O, Clayton R, Kirkness E, Kerlavage A, Bult C, Tomb J, Dougherty B, Merrick J, McKenney K, Sutton G, Fitzhugh W, Fields C, Gocyne J, Scott J, Shirley R, Liu L, Glodek A, Kelley J, Jenny M, Weidman J, Phillips C, Spriggs T, Hedblom E, Cotton M, Utterback T, Hanna M, Nguyen D, Saudek D, Brandon R, Fine L, Fritchman J, Fuhrmann J, Geoghagen N, Gnehm C, McDonald L, Small K, Fraser C, Smith H, Venter J. Whole-genome Random Sequencing and Assembly of Haemophilus influenzae Rd. Science. 1995;269(5223):496–512. doi: 10.1126/science.7542800. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

